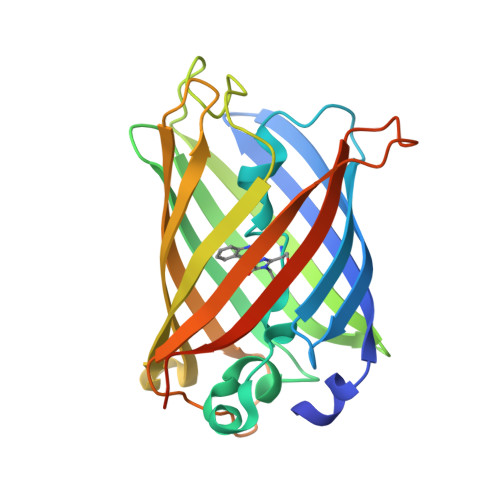
Structure of a Fluorescent Protein from Aequorea Victoria Bearing the Obligate-Monomer Mutation A206K.
von Stetten, D., Noirclerc-Savoye, M., Goedhart, J., Gadella, T.W.J.J., Royant, A.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 878
- PubMed: 22869113
- DOI: https://doi.org/10.1107/S1744309112028667
- Primary Citation of Related Structures:
4AR7 - PubMed Abstract:
The green fluorescent protein (GFP) from the jellyfish Aequoria victoria has been shown to dimerize at high concentrations, which could lead to artefacts in imaging experiments. To ensure a truly monomeric state, an A206K mutation has been introduced into most of its widely used variants, with minimal effect on the spectroscopic properties. Here, the first structure of one of these variants, the cyan fluorescent protein mTurquoise, is presented and compared with that of its dimeric version mTurquoise-K206A. No significant structural change is detected in the chromophore cavity, reinforcing the notion that this mutation is spectroscopically silent and validating that the structural analysis performed on dimeric mutants also applies to monomeric versions. Finally, it is explained why cyan versions of GFP containing the Y66W and N146I mutations do not require the A206K mutation to prevent dimerization at high concentrations.
- Structural Biology Group, European Synchrotron Radiation Facility, 6 Rue Jules Horowitz, 38043 Grenoble, France.
Organizational Affiliation: