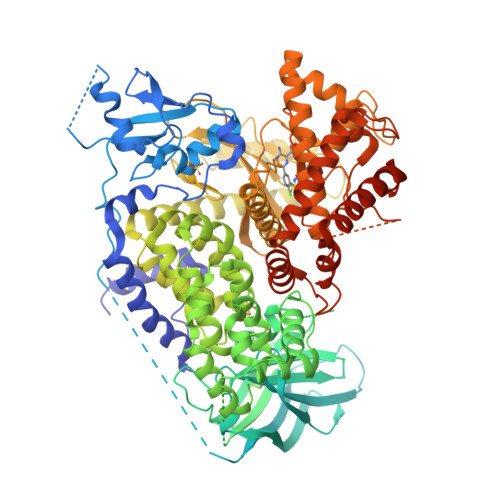
The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
Leahy, J.W., Buhr, C.A., Johnson, H.W.B., Kim, B.G., Baik, T., Cannoy, J., Forsyth, T.P., Jeong, J.W., Lee, M.S., Ma, S., Noson, K., Wang, L., Williams, M., Nuss, J.M., Brooks, E., Heald, N., Holst, C., Jaeger, C., Lam, S., Lougheed, J.C., Nguyen, L., Plonowski, A., Stout, T., Foster, P.G., Wu, X., Yakes, M.F., Yu, R., Zhang, W., Lamb, P., Raeber, O.(2012) J Med Chem 55: 5467
- PubMed: 22548342
- DOI: https://doi.org/10.1021/jm300403a
- Primary Citation of Related Structures:
4ANU, 4ANV, 4ANW, 4ANX - PubMed Abstract:
The phosphoinositide 3-kinases (PI3Ks) have been linked to an extraordinarily diversified group of cellular functions making these enzymes compelling targets for the treatment of disease. A large body of evidence has linked PI3Kγ to the modulation of autoimmune and inflammatory processes making it an intriguing target for drug discovery. Our high-throughput screening (HTS) campaign revealed two hits that were nominated for further optimization studies. The in vitro activity of the first HTS hit, designated as the sulfonylpiperazine scaffold, was optimized utilizing structure-based design. However, nonoptimal pharmacokinetic properties precluded this series from further studies. An overlay of the X-ray structures of the sulfonylpiperazine scaffold and the second HTS hit within their complexes with PI3Kγ revealed a high degree of overlap. This feature was utilized to design a series of hybrid analogues including advanced leads such as 31 with desirable potency, selectivity, and oral bioavailability.
Organizational Affiliation:
Department of Drug Discovery, Exelixis, 169 Harbor Way, South San Francisco, California 94083, United States.