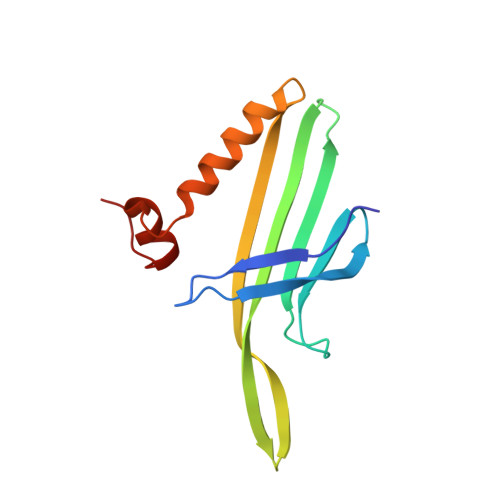
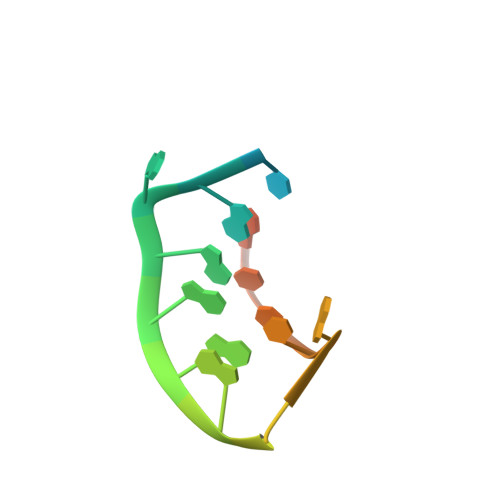
Prr1 Coat Protein Binding to its RNA Translational Operator
Persson, M., Tars, K., Liljas, L.(2013) Acta Crystallogr D Biol Crystallogr 69: 367
- PubMed: 23519411
- DOI: https://doi.org/10.1107/S0907444912047464
- Primary Citation of Related Structures:
4ANG - PubMed Abstract:
In small RNA bacteriophages, the genomic RNA binds to the coat proteins when the viral capsid assembles. This is achieved through sequence-specific interactions between a coat-protein dimer and an RNA stem-loop that includes the start codon for the replicase gene. The structure of virus-like particles of the small RNA phage PRR1 bound to an RNA segment corresponding to this stem-loop has been solved and the binding was compared with the related, and better investigated, phage MS2. The overall conformation of the RNA is found to be similar and the residues that are involved in RNA binding in PRR1 are the same as in MS2. The arrangement of the nucleotide bases in the loop of the stem-loop is different, leading to a difference in the stacking at the conserved Tyr86, which is equivalent to Tyr85 in MS2.
Organizational Affiliation:
Department of Cell and Molecular Biology, Uppsala University, BMC, Husargatan 3, Box 596, S-751 24 Uppsala, Sweden.