Curing a Mutant Transcarbamylase with a Small Molecule: A Crystallographic View
Polo, L.M., Rubio, V.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PUTRESCINE CARBAMOYLTRANSFERASE | 355 | Enterococcus faecalis | Mutation(s): 1 EC: 2.1.3.6 | 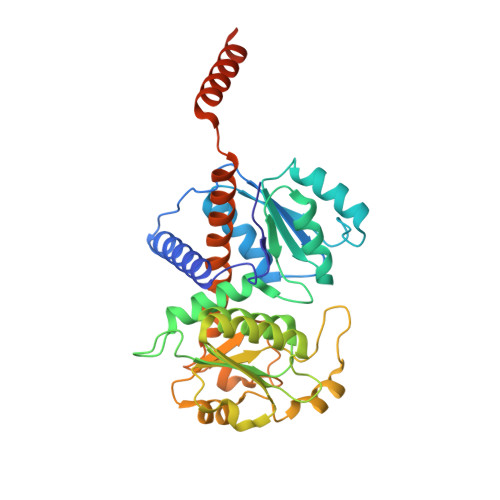 | |
UniProt | |||||
Find proteins for Q837U7 (Enterococcus faecalis (strain ATCC 700802 / V583)) Explore Q837U7 Go to UniProtKB: Q837U7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q837U7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PAO Query on PAO | AA [auth F] H [auth A] M [auth B] Q [auth C] T [auth D] | N-(PHOSPHONOACETYL)-L-ORNITHINE C7 H15 N2 O6 P FCIHAQFHXJOLIF-YFKPBYRVSA-N |  | ||
| GLN Query on GLN | G [auth A], P [auth C], W [auth E] | GLUTAMINE C5 H10 N2 O3 ZDXPYRJPNDTMRX-VKHMYHEASA-N |  | ||
| ILE Query on ILE | L [auth B] | ISOLEUCINE C6 H13 N O2 AGPKZVBTJJNPAG-WHFBIAKZSA-N |  | ||
| TRS Query on TRS | CA [auth F], J [auth A], O [auth B], S [auth C] | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C4 H12 N O3 LENZDBCJOHFCAS-UHFFFAOYSA-O |  | ||
| GAI Query on GAI | BA [auth F] I [auth A] N [auth B] R [auth C] U [auth D] | GUANIDINE C H5 N3 ZRALSGWEFCBTJO-UHFFFAOYSA-N |  | ||
| NI Query on NI | K [auth A], Z [auth F] | NICKEL (II) ION Ni VEQPNABPJHWNSG-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 103.69 | α = 90 |
| b = 130.6 | β = 104.25 |
| c = 164.157 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |
| MOLREP | phasing |