Peptide Linkage to the Alpha-Subunit of Mhcii Creates a Stably Inverted Antigen Presentation Complex.
Schlundt, A., Gunther, S., Sticht, J., Wieczorek, M., Roske, Y., Heinemann, U., Freund, C.(2012) J Mol Biology 423: 294
- PubMed: 22820093
- DOI: https://doi.org/10.1016/j.jmb.2012.07.008
- Primary Citation of Related Structures:
4AEN, 4AH2 - PubMed Abstract:
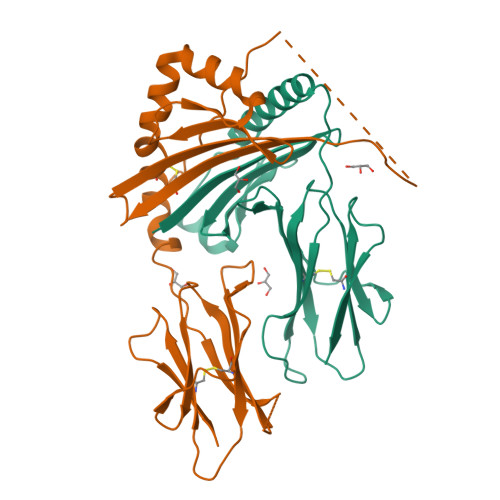
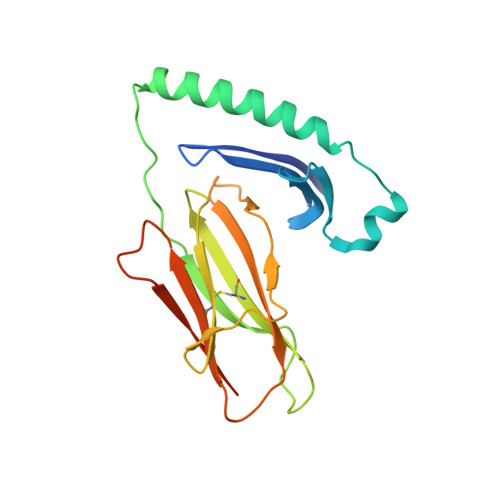
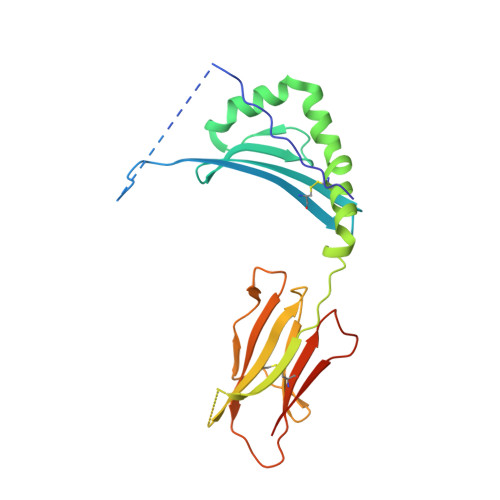
Class II proteins of the major histocompatibility complex (MHCII) typically present exogenous antigenic peptides to cognate T cell receptors of CD4-T lymphocytes. The exact conformation of peptide-MHCII complexes (pMHCII) can vary depending on the length, register and orientation of the bound peptide. We have recently found the self-peptide CLIP (class-II-associated invariant chain-derived peptide) to adopt a dynamic bidirectional binding mode with regard to the human MHCII HLA-DR1 (HLA, human leukocyte antigen). We suggested that inversely bound peptides could activate specific T cell clones in the context of autoimmunity. As a first step to prove this hypothesis, pMHC complexes restricted to either the canonical or the inverted peptide orientation have to be constructed. Here, we show that genetically encoded linkage of CLIP and two other antigenic peptides to the HLA-DR1 α-chain results in stable complexes with inversely bound ligands. Two-dimensional NMR and biophysical analyses indicate that the CLIP-bound pMHC(inv) complex (pMHC(inv), inverted MHCII-peptide complex) displays high thermodynamic stability but still allows for the exchange against higher-affinity viral antigen. Complemented by comparable data on a corresponding β-chain-fused canonical HLA-DR1/CLIP complex, we further show that linkage of CLIP leads to a binding mode exactly the same as that of the corresponding unlinked constructs. We suggest that our approach constitutes a general strategy to create pMHC(inv) complexes. Such engineering is needed to create orientation-specific antibodies and raise T cells to study phenomena of autoimmunity caused by isomeric pMHCs.
Organizational Affiliation:
Protein Biochemistry Group, Freie Universität Berlin, Thielallee 63, 14195 Berlin, Germany.