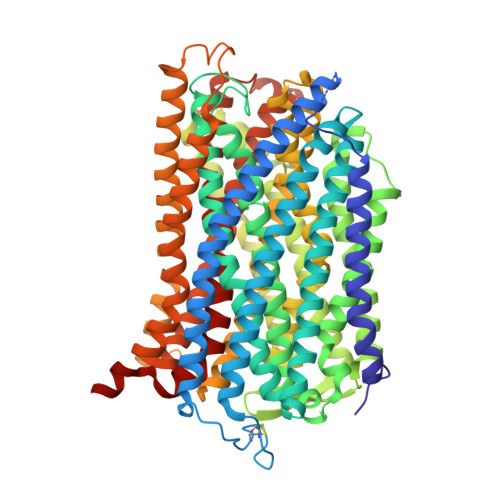
Crystal Structure of a Membrane Embedded H1-Translocating Pyrophosphatase
Lin, S.-M., Tsai, J.-Y., Hsiao, C.-D., Chiu, C.-L., Pan, R.-L., Sun, Y.-J.(2012) Nature 484: 399
- PubMed: 22456709
- DOI: https://doi.org/10.1038/nature10963
- Primary Citation of Related Structures:
4A01 - PubMed Abstract:
H(+)-translocating pyrophosphatases (H(+)-PPases) are active proton transporters that establish a proton gradient across the endomembrane by means of pyrophosphate (PP(i)) hydrolysis. H(+)-PPases are found primarily as homodimers in the vacuolar membrane of plants and the plasma membrane of several protozoa and prokaryotes. The three-dimensional structure and detailed mechanisms underlying the enzymatic and proton translocation reactions of H(+)-PPases are unclear. Here we report the crystal structure of a Vigna radiata H(+)-PPase (VrH(+)-PPase) in complex with a non-hydrolysable substrate analogue, imidodiphosphate (IDP), at 2.35 Å resolution. Each VrH(+)-PPase subunit consists of an integral membrane domain formed by 16 transmembrane helices. IDP is bound in the cytosolic region of each subunit and trapped by numerous charged residues and five Mg(2+) ions. A previously undescribed proton translocation pathway is formed by six core transmembrane helices. Proton pumping can be initialized by PP(i) hydrolysis, and H(+) is then transported into the vacuolar lumen through a pathway consisting of Arg 242, Asp 294, Lys 742 and Glu 301. We propose a working model of the mechanism for the coupling between proton pumping and PP(i) hydrolysis by H(+)-PPases.
Organizational Affiliation:
Department of Life Science and Institute of Bioinformatics and Structural Biology, College of Life Science, National Tsing Hua University, Hsinchu 30013, Taiwan.