Structure of an RNA duplex with an unusual G.C pair in wobble-like conformation at 1.6 A resolution.
Perbandt, M., Vallazza, M., Lippmann, C., Betzel, C., Erdmann, V.A.(2001) Acta Crystallogr D Biol Crystallogr 57: 219-224
- PubMed: 11173467
- DOI: https://doi.org/10.1107/s0907444900017042
- Primary Citation of Related Structures:
439D - PubMed Abstract:
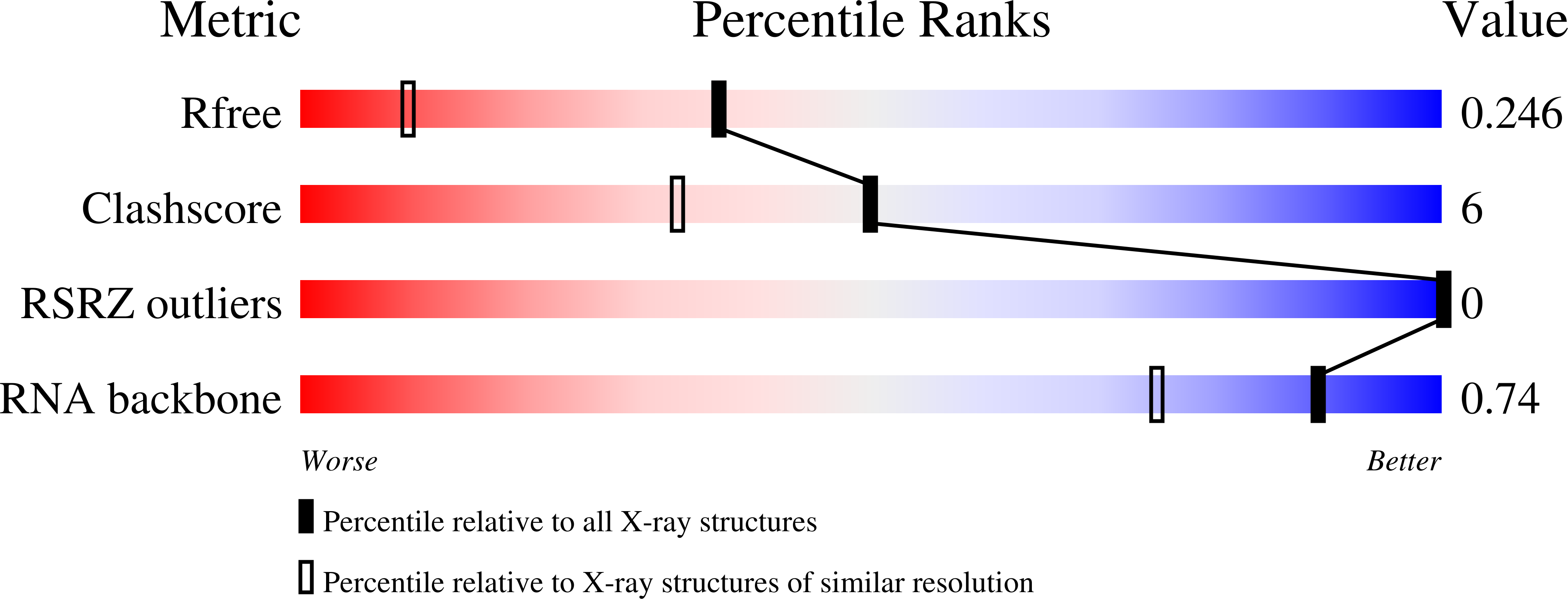
The structure of the RNA duplex r(CUGGGCGG).r(CCGCCUGG) has been determined at 1.6 A resolution and refined to a final R factor of 18.3% (R(free) = 24.1%). The sequence of the RNA fragment resembles domain E of Thermus flavus 5S rRNA. A previously undescribed wobble-like G.C base-pair formation is found. Owing to the observed hydrogen-bond network, it is proposed that the cytosine is protonated at position N3. The unusual base-pair formation is presumably strained by intermolecular interactions. In this context, crystal packing and particular intermolecular contacts may have direct influence on the three-dimensional structure. Furthermore, this structure includes two G.U wobble base pairs in tandem conformation, with the purines forming a so-called 'cross-strand G stack'.
Organizational Affiliation:
Institute of Medical Biochemistry and Molecular Biology, University Hospital Eppendorf, c/o DESY Building 22a, Notkestrasse 85, 22603 Hamburg, Germany.