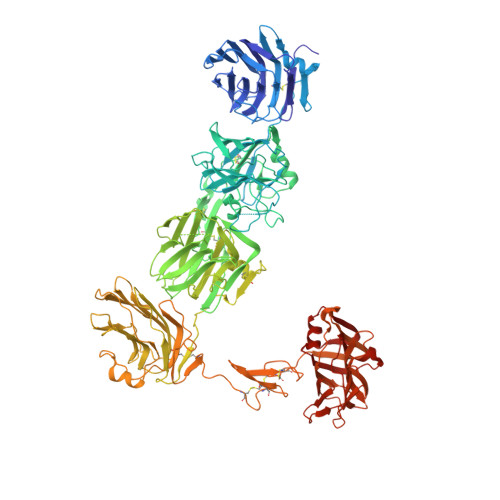
The crystal structure of the alpha-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function.
Miller, M.T., Mileni, M., Comoletti, D., Stevens, R.C., Harel, M., Taylor, P.(2011) Structure 19: 767-778
- PubMed: 21620717
- DOI: https://doi.org/10.1016/j.str.2011.03.011
- Primary Citation of Related Structures:
3POY - PubMed Abstract:
α- and β-neurexins (NRXNs) are transmembrane cell adhesion proteins that localize to presynaptic membranes in neurons and interact with the postsynaptic neuroligins (NLGNs). Their gene mutations are associated with the autism spectrum disorders. The extracellular region of α-NRXNs, containing nine independently folded domains, has structural complexity and unique functional characteristics, distinguishing it from the smaller β-NRXNs. We have solved the X-ray crystal structure of seven contiguous domains of the α-NRXN-1 extracellular region at 3.0 Å resolution. The structure reveals an arrangement where the N-terminal five domains adopt a more rigid linear conformation and the two C-terminal domains form a separate arm connected by a flexible hinge. In an extended conformation the molecule is suitably configured to accommodate a bound NLGN molecule, as supported by structural comparison and surface plasmon resonance. These studies provide the structural basis for a multifunctional synaptic adhesion complex mediated by α-NRXN-1.
- Department of Pharmacology, Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, La Jolla, CA 92093, USA. m4miller@ucsd.edu
Organizational Affiliation: