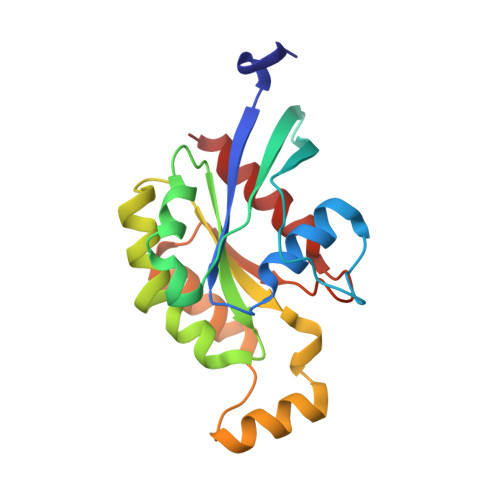
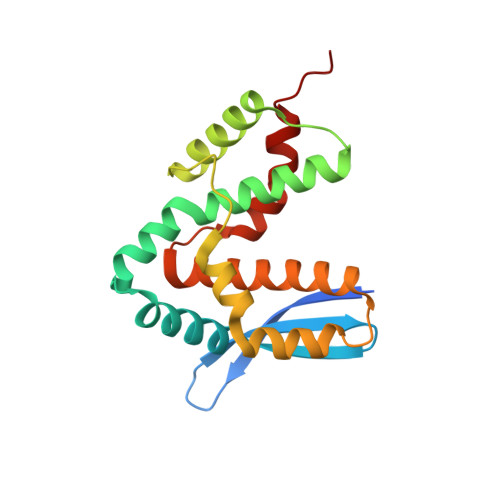
Structure of Shigella IPGB2 in complex with human RhoA: Implications for the mechanism of bacterial GEF-mimicry
Klink, B.U., Barden, S., Heidler, T.V., Borchers, C., Ladwein, M., Stradal, T.E.B., Rottner, K., Heinz, D.W.(2010) J Biological Chem 285: 17197-17208
- PubMed: 20363740
- DOI: https://doi.org/10.1074/jbc.M110.107953
- Primary Citation of Related Structures:
3LW8, 3LWN, 3LXR, 3LYQ - PubMed Abstract:
A common theme in bacterial pathogenesis is the manipulation of eukaryotic cells by targeting the cytoskeleton. This is in most cases achieved either by modifying actin, or indirectly via activation of key regulators controlling actin dynamics such as Rho-GTPases. A novel group of bacterial virulence factors termed the WXXXE family has emerged as guanine nucleotide exchange factors (GEFs) for these GTPases. The precise mechanism of nucleotide exchange, however, has remained unclear. Here we report the structure of the WXXXE-protein IpgB2 from Shigella flexneri and its complex with human RhoA. We unambiguously identify IpgB2 as a bacterial RhoA-GEF and dissect the molecular mechanism of GDP release, an essential prerequisite for GTP binding. Our observations uncover that IpgB2 induces conformational changes on RhoA mimicking DbI- but not DOCK family GEFs. We also show that dissociation of the GDP.Mg(2+) complex is preceded by the displacement of the metal ion to the alpha-phosphate of the nucleotide, diminishing its affinity to the GTPase. These data refine our understanding of the mode of action not only of WXXXE GEFs but also of mammalian GEFs of the DH/PH family.
- Division of Structural Biology, Helmholtz Zentrum für Infektionsforschung, D-38124 Braunschweig, Germany.
Organizational Affiliation: