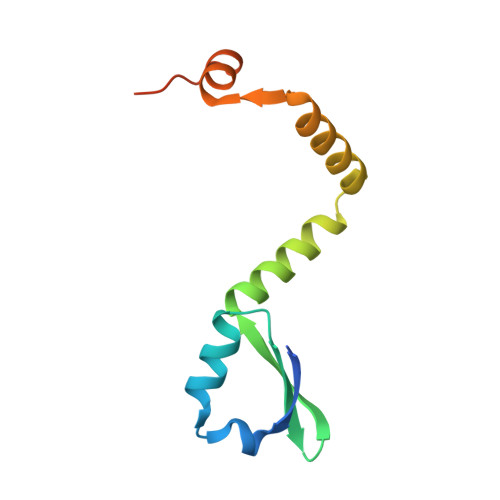
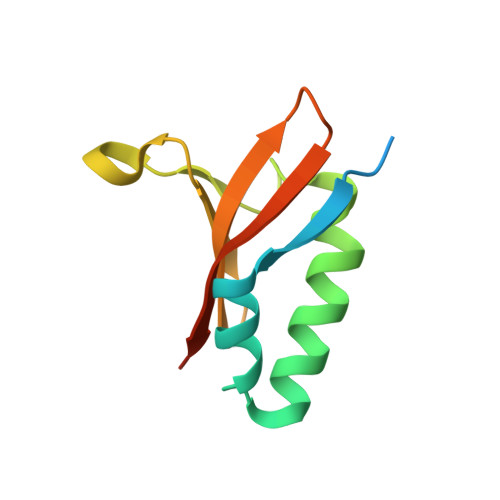
Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes.
Miallau, L., Jain, P., Arbing, M.A., Cascio, D., Phan, T., Ahn, C.J., Chan, S., Chernishof, I., Maxson, M., Chiang, J., Jacobs Jr., W.R., Eisenberg, D.S.(2013) Structure 21: 627-637
- PubMed: 23523424
- DOI: https://doi.org/10.1016/j.str.2013.02.008
- Primary Citation of Related Structures:
3G5O, 3OEI - PubMed Abstract:
The Mycobacterium tuberculosis (Mtb) genome encodes approximately 90 toxin-antitoxin protein complexes, including three RelBE family members, which are believed to play a major role in bacterial fitness and pathogenicity. We have determined the crystal structures of Mtb RelBE-2 and RelBE-3, and the structures reveal homologous heterotetramers. Our structures suggest RelE-2, and by extension the closely related RelE-1, use a different catalytic mechanism than RelE-3, because our analysis of the RelE-2 structure predicts additional amino acid residues that are likely to be functionally significant and are missing from analogous positions in the RelE-3 structure. Toxicity assays corroborate our structural findings; overexpression of RelE-3, whose active site is more similar to Escherichia coli YoeB, has limited consequences on bacterial growth, whereas RelE-1 and RelE-2 overexpression results in acute toxicity. Moreover, RelE-2 overexpression results in an elongated cell phenotype in Mycobacterium smegmatis and protects M. tuberculosis against antibiotics, suggesting a different functional role for RelE-2.
- Howard Hughes Medical Institute and UCLA-DOE Institute for Genomics and Proteomics and Department of Biological Chemistry, University of California, Los Angeles, Los Angeles, CA 90095-1570, USA.
Organizational Affiliation: