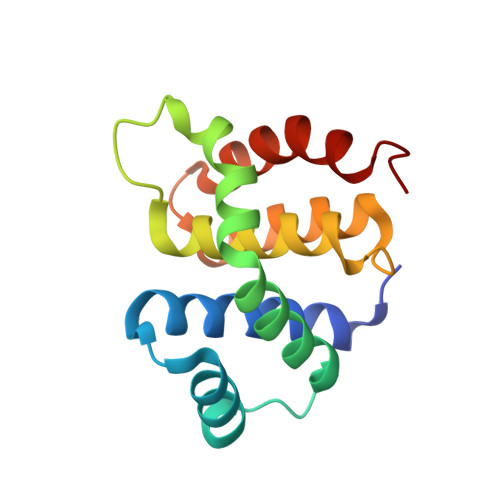
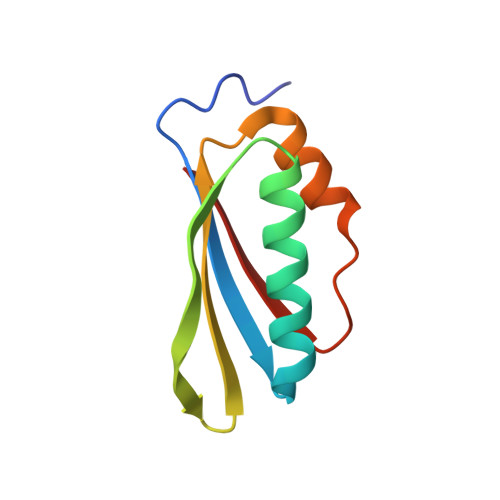
Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex.
Luo, X., Hsiao, H.H., Bubunenko, M., Weber, G., Court, D.L., Gottesman, M.E., Urlaub, H., Wahl, M.C.(2008) Mol Cell 32: 791-802
- PubMed: 19111659
- DOI: https://doi.org/10.1016/j.molcel.2008.10.028
- Primary Citation of Related Structures:
3D3B, 3D3C - PubMed Abstract:
Protein S10 is a component of the 30S ribosomal subunit and participates together with NusB protein in processive transcription antitermination. The molecular mechanisms by which S10 can act as a translation or a transcription factor are not understood. We used complementation assays and recombineering to delineate regions of S10 dispensable for antitermination, and determined the crystal structure of a transcriptionally active NusB-S10 complex. In this complex, S10 adopts the same fold as in the 30S subunit and is blocked from simultaneous association with the ribosome. Mass spectrometric mapping of UV-induced crosslinks revealed that the NusB-S10 complex presents an intermolecular, composite, and contiguous binding surface for RNAs containing BoxA antitermination signals. Furthermore, S10 overproduction complemented a nusB null phenotype. These data demonstrate that S10 and NusB together form a BoxA-binding module, that NusB facilitates entry of S10 into the transcription machinery, and that S10 represents a central hub in processive antitermination.
- Research Group X-Ray Crystallography, Max-Planck-Institute for Biophysical Chemistry, D-37077 Göttingen, Germany.
Organizational Affiliation: