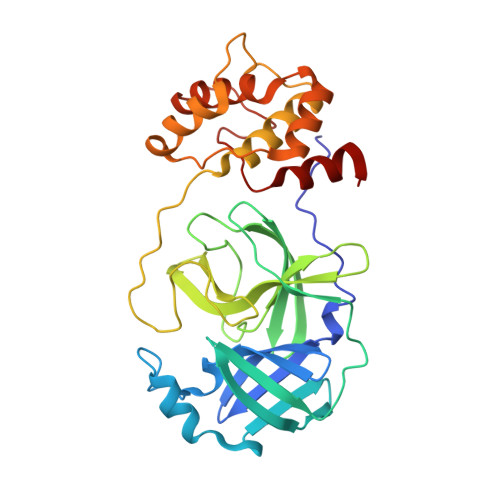
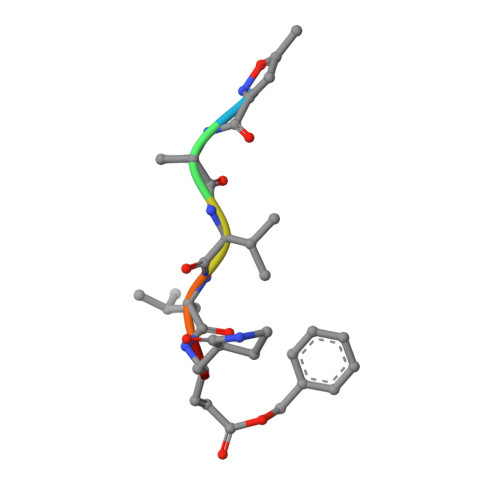
Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1.
Zhao, Q., Li, S., Xue, F., Zou, Y., Chen, C., Bartlam, M., Rao, Z.(2008) J Virol 82: 8647-8655
- PubMed: 18562531
- DOI: https://doi.org/10.1128/JVI.00298-08
- Primary Citation of Related Structures:
3D23 - PubMed Abstract:
The newly emergent human coronavirus HKU1 (HCoV-HKU1) was first identified in Hong Kong in 2005. Infection by HCoV-HKU1 occurs worldwide and causes syndromes such as the common cold, bronchitis, and pneumonia. The CoV main protease (M(pro)), which is a key enzyme in viral replication via the proteolytic processing of the replicase polyproteins, has been recognized as an attractive target for rational drug design. In this study, we report the structure of HCoV-HKU1 M(pro) in complex with a Michael acceptor, inhibitor N3. The structure of HCoV-HKU1 provides a high-quality model for group 2A CoVs, which are distinct from group 2B CoVs such as severe acute respiratory syndrome CoV. The structure, together with activity assays, supports the relative conservation at the P1 position that was discovered by sequencing the HCoV-HKU1 genome. Combined with structural data from other CoV M(pro)s, the HCoV-HKU1 M(pro) structure reported here provides insights into both substrate preference and the design of antivirals targeting CoVs.
- Tsinghua-Nankai-IBP Joint Research Group for Structural Biology, Tsinghua University, Beijing, China.
Organizational Affiliation: