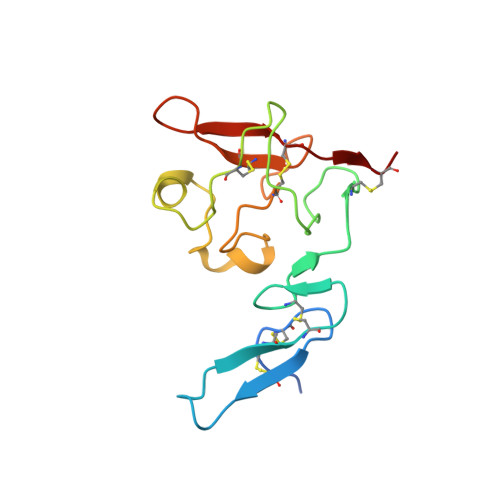
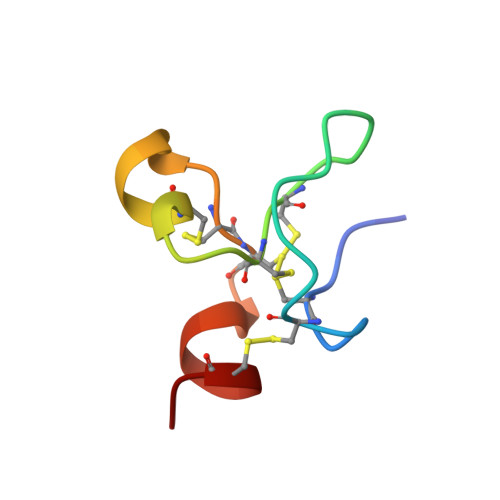
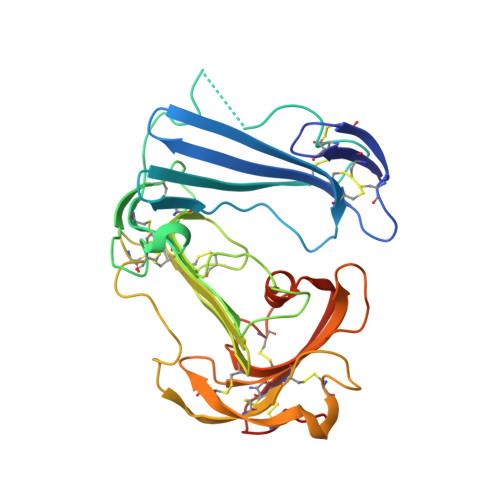
Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes
Huai, Q., Zhou, A., Lin, L., Mazar, A.P., Parry, G.C., Callahan, J., Shaw, D.E., Furie, B., Furie, B.C., Huang, M.(2008) Nat Struct Mol Biol 15: 422-423
- PubMed: 18376415
- DOI: https://doi.org/10.1038/nsmb.1404
- Primary Citation of Related Structures:
3BT1, 3BT2 - PubMed Abstract:
The urokinase receptor (uPAR) can recognize several ligands. The structural basis for this multiple ligand recognition by uPAR is unknown. This study reports the crystal structures of uPAR in complex with both urokinase (uPA) and vitronectin and reveal that uPA occupies the central cavity of the receptor, whereas vitronectin binds at the outer side of the receptor. These results provide a structural understanding of one receptor binding to two ligands.
- Division of Hemostasis and Thrombosis, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, Massachusetts 02215, USA.
Organizational Affiliation: