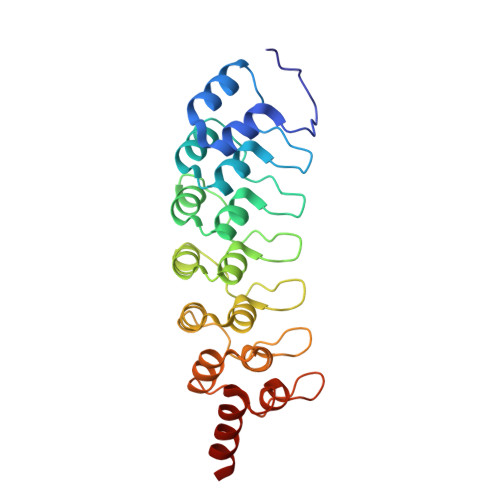
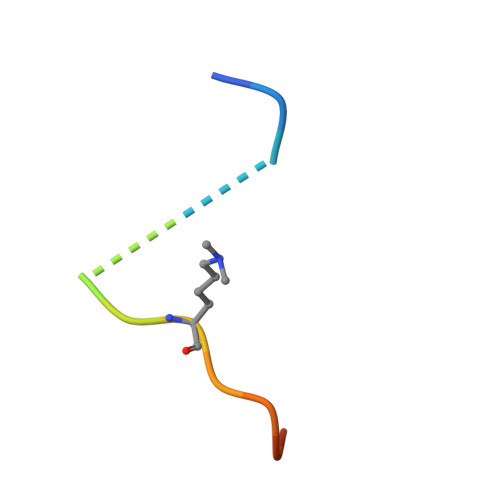
The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules.
Collins, R.E., Northrop, J.P., Horton, J.R., Lee, D.Y., Zhang, X., Stallcup, M.R., Cheng, X.(2008) Nat Struct Mol Biol 15: 245-250
- PubMed: 18264113
- DOI: https://doi.org/10.1038/nsmb.1384
- Primary Citation of Related Structures:
3B7B, 3B95 - PubMed Abstract:
Histone modifications have important roles in transcriptional control, mitosis and heterochromatin formation. G9a and G9a-like protein (GLP) are euchromatin-associated methyltransferases that repress transcription by mono- and dimethylating histone H3 at Lys9 (H3K9). Here we demonstrate that the ankyrin repeat domains of G9a and GLP bind with strong preference to N-terminal H3 peptides containing mono- or dimethyl K9. X-ray crystallography revealed the basis for recognition of the methylated lysine by a partial hydrophobic cage with three tryptophans and one acidic residue. Substitution of key residues in the cage eliminated the H3 tail interaction. Hence, G9a and GLP contain a new type of methyllysine binding module (the ankyrin repeat domains) and are the first examples of protein (histone) methyltransferases harboring in a single polypeptide the activities that generate and read the same epigenetic mark.
- Department of Biochemistry, Emory University School of Medicine, 1510 Clifton Road, Atlanta, Georgia 30322, USA.
Organizational Affiliation: