Moonlighting by Different Stressors: Crystal Structure of the Chaperone Species of a 2-Cys Peroxiredoxin.
Saccoccia, F., Di Micco, P., Boumis, G., Brunori, M., Koutris, I., Miele, A.E., Morea, V., Sriratana, P., Williams, D.L., Bellelli, A., Angelucci, F.(2012) Structure 20: 429
- PubMed: 22405002
- DOI: https://doi.org/10.1016/j.str.2012.01.004
- Primary Citation of Related Structures:
3ZTL, 3ZVJ - PubMed Abstract:
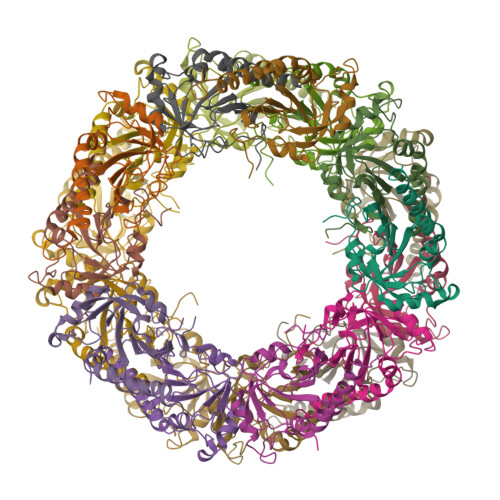
2-Cys peroxiredoxins (Prxs) play two different roles depending on the physiological status of the cell. They are thioredoxin-dependent peroxidases under low oxidative stress and ATP-independent chaperones upon exposure to high peroxide concentrations. These alternative functions have been associated with changes in the oligomerization state from low-(LMW) to high-molecular-weight (HMW) species. Here we present the structures of Schistosoma mansoni PrxI in both states: the LMW decamer and the HMW 20-mer formed by two stacked decamers. The latter is the structure of a 2-Cys Prx chaperonic form. Comparison of the structures sheds light on the mechanism by which chemical stressors, such as high H(2)O(2) concentration and acidic pH, are sensed and translated into a functional switch in this protein family. We also propose a model to account for the in vivo formation of long filaments of stacked Prx rings.
Organizational Affiliation:
Department of Biochemical Sciences, Sapienza University of Rome and Istituto Pasteur-Fondazione Cenci Bolognetti, P.le Aldo Moro 5, 00185 Rome, Italy.