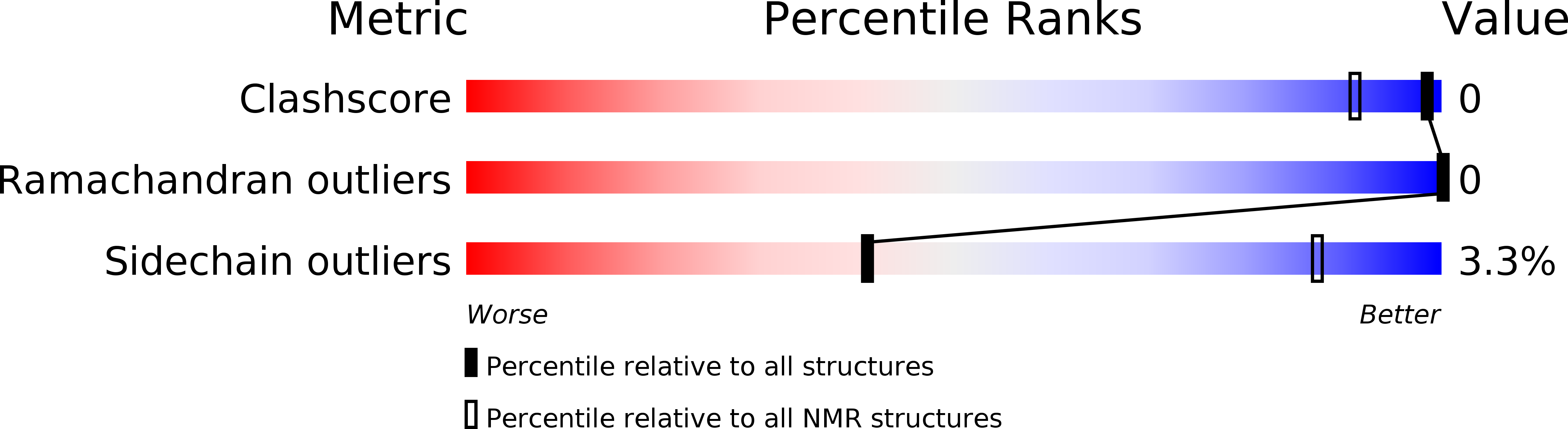Investigation of Homeodomain Membrane Translocation Properties: Insights from the Structure Determination of Engrailed-2 Homeodomain in Aqueous and Membrane-Mimetic Environments.
Carlier, L., Balayssac, S., Cantrelle, F., Khemtemourian, L., Chassaing, G., Joliot, A., Lequin, O.(2013) Biophys J 105: 667
- PubMed: 23931315
- DOI: https://doi.org/10.1016/j.bpj.2013.06.024
- Primary Citation of Related Structures:
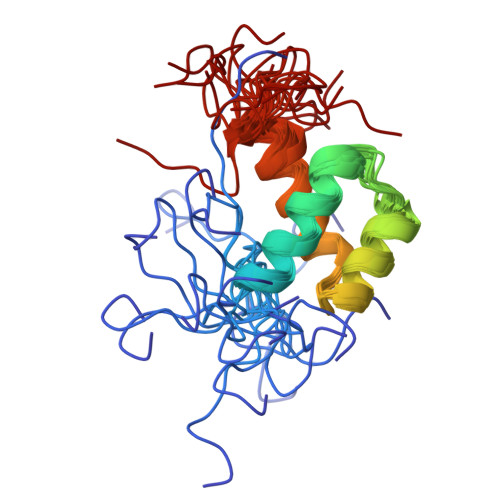
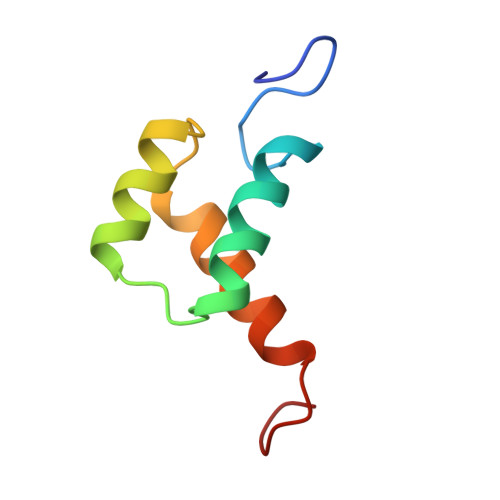
3ZOB - PubMed Abstract:
In addition to their well-known DNA-binding properties, homeodomains have the ability to efficiently translocate across biological membranes through still poorly-characterized mechanisms. To date, most biophysical studies addressing the mechanisms of internalization have focused on small synthetic peptides rather than full-length globular homeodomains. In this work, we characterized the conformational properties of chicken Engrailed 2 homeodomain (En2HD) in aqueous solution and in membrane mimetic environments using circular dichroism, Trp fluorescence, and NMR spectroscopy. En2HD adopts a well-defined three-helical bundle fold in aqueous solution. The Trp-48 residue, which is critical for internalization, is fully buried in the hydrophobic core. Circular dichroism and fluorescence reveal that a conformational transition occurs in anionic lipid vesicles and in micelles. En2HD loses its native three-dimensional structure in micellar environments but, remarkably, near-native helical secondary structures are maintained. Long-range interactions could be detected using site-directed spin labels, indicating that the three helices do not adopt extended orientations. Noncovalent paramagnetic probes yielded information about helix positioning and unveiled the burial of critical aromatic and basic residues within the micelles. Our results suggest that electrostatic interactions with membranes may be determinant in inducing a conformational change enabling Trp-48 to insert into membranes.
Organizational Affiliation:
Université Pierre et Marie Curie, Centre National de la Recherche Scientifique, Ecole Normale Supérieure, Laboratoire des Biomolécules, UMR, Paris, France.