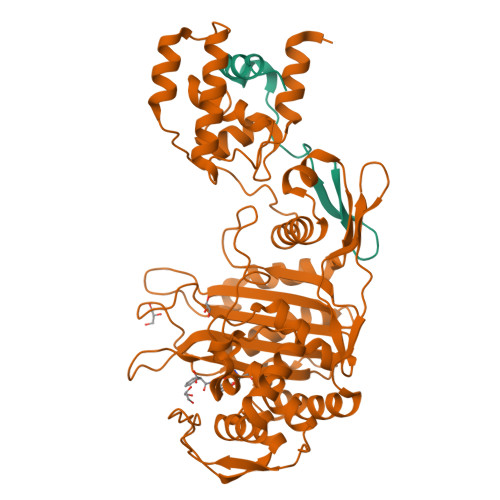
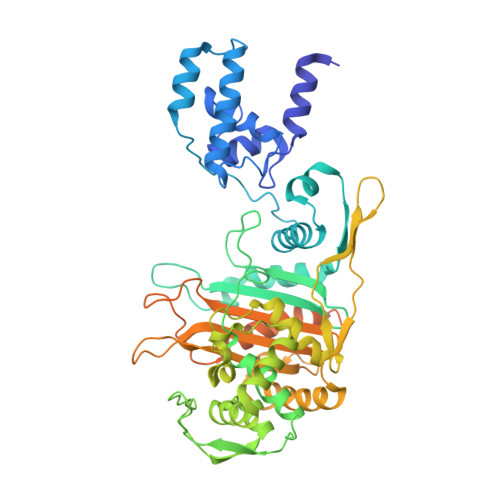
Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Jeong, J., Kim, Y., Rojviriya, C., Ha, S., Kang, B.S., Kim, Y.(2013) Antimicrob Agents Chemother 57: 3507
- PubMed: 23669378
- DOI: https://doi.org/10.1128/AAC.00144-13
- Primary Citation of Related Structures:
3ZG7, 3ZG8, 3ZGA - PubMed Abstract:
Penicillin-binding proteins (PBPs), which catalyze the biosynthesis of the peptidoglycan chain of the bacterial cell wall, are the major molecular target of bacterial antibiotics. Here, we present the crystal structures of the bifunctional peptidoglycan glycosyltransferase (GT)/transpeptidase (TP) PBP4 from Listeria monocytogenes in the apo-form and covalently linked to two β-lactam antibiotics, ampicillin and carbenicillin. The orientation of the TP domain with respect to the GT domain is distinct from that observed in the previously reported structures of bifunctional PBPs, suggesting interdomain flexibility. In this structure, the active site of the GT domain is occluded by the close apposition of the linker domain, which supports the hypothesis that interdomain flexibility is related to the regulation of GT activity. The acylated structures reveal the mode of action of β-lactam antibiotics toward the class A PBP4 from the human pathogen L. monocytogenes. Ampicillin and carbenicillin can access the active site and be acylated without requiring a structural rearrangement. In addition, the active site of the TP domain in the apo-form is occupied by the tartrate molecule via extensive hydrogen bond interactions with the catalytically important residues; thus, derivatives of the tartrate molecule may be useful in the search for new antibiotics to inhibit PBPs.
Organizational Affiliation:
Pohang Accelerator Laboratory, Pohang University of Science and Technology, Pohang, Republic of Korea.