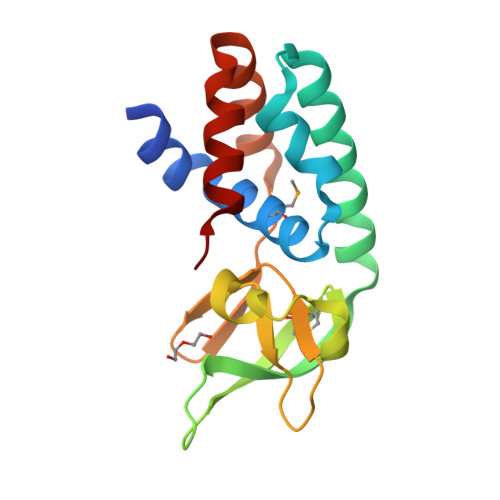
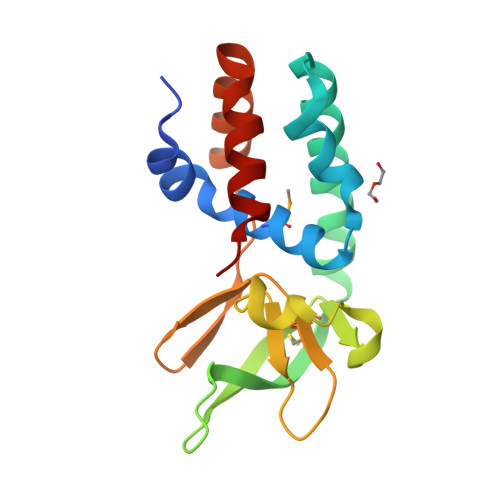
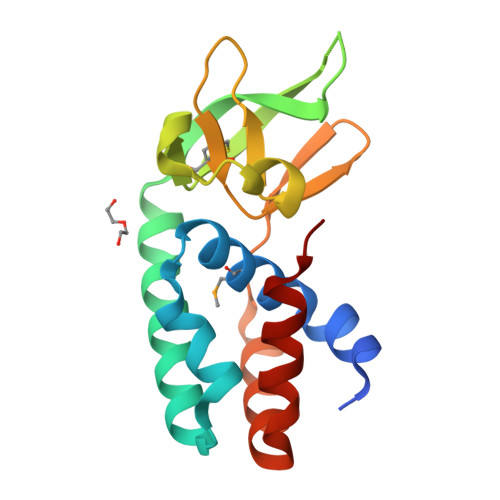
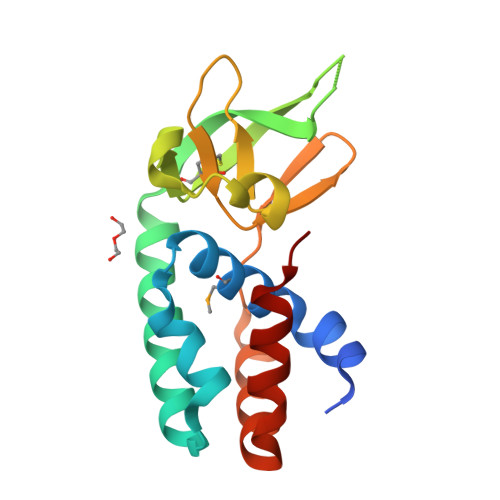
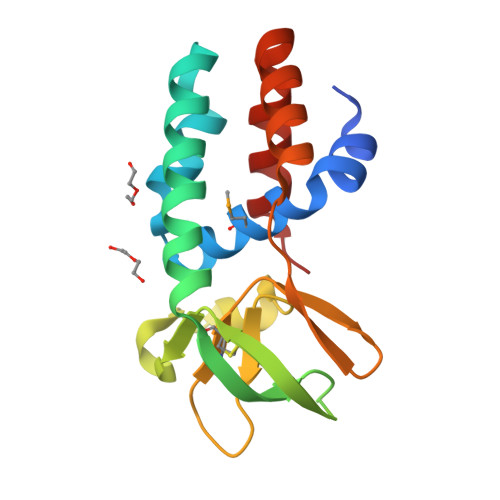
Structure of the Archaeal Cascade Subunit Csa5: Relating the Small Subunits of Crispr Effector Complexes.
Reeks, J., Graham, S., Anderson, L., Liu, H., White, M.F., Naismith, J.H.(2013) RNA Biol 10: 762
- PubMed: 23846216
- DOI: https://doi.org/10.4161/rna.23854
- Primary Citation of Related Structures:
3ZC4 - PubMed Abstract:
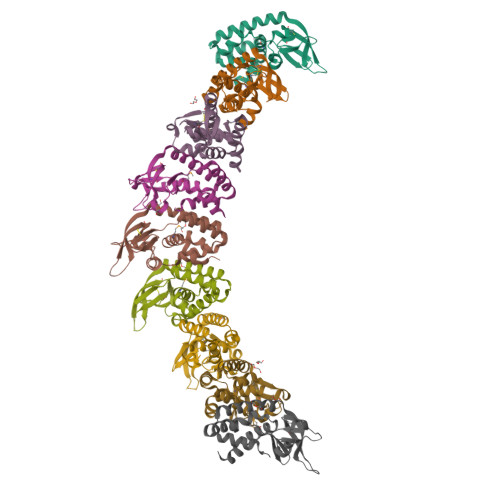
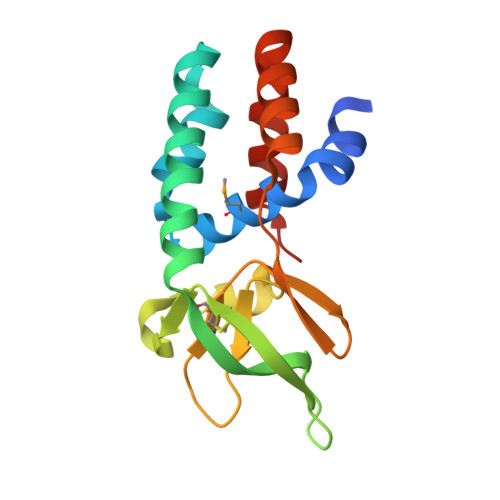
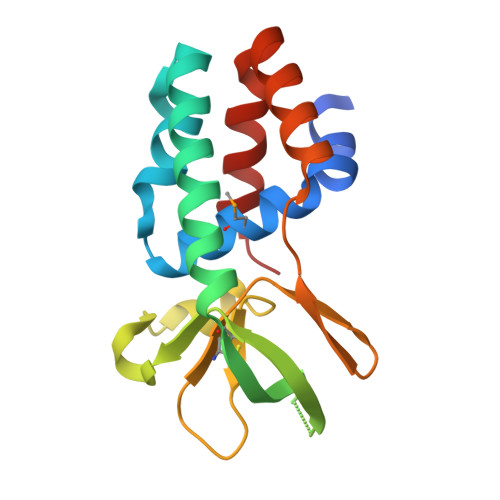
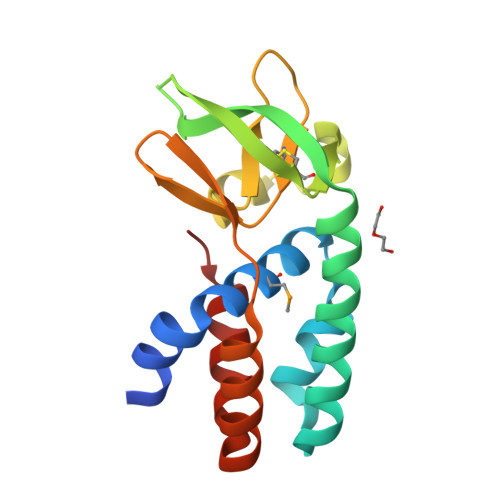
The Cascade complex for CRISPR-mediated antiviral immunity uses CRISPR RNA (crRNA) to target invading DNA species from mobile elements such as viruses, leading to their destruction. The core of the Cascade effector complex consists of the Cas5 and Cas7 subunits, which are widely conserved in prokaryotes. Cas7 binds crRNA and forms the helical backbone of Cascade. Many archaea encode a version of the Cascade complex (denoted Type I-A) that includes a Csa5 (or small) subunit, which interacts weakly with the core proteins. Here, we report the crystal structure of the Csa5 protein from Sulfolobus solfataricus. Csa5 comprises a conserved α-helical domain with a small insertion consisting of a weakly conserved β-strand domain. In the crystal, the Csa5 monomers have multimerized into infinite helical threads. At each interface is a strictly conserved intersubunit salt bridge, deletion of which disrupts multimerization. Structural analysis indicates a shared evolutionary history among the small subunits of the CRISPR effector complexes. The same α-helical domain is found in the C-terminal domain of Cse2 (from Type I-E Cascade), while the N-terminal domain of Cse2 is found in Cmr5 of the CMR (Type III-B) effector complex. As Cmr5 shares no match with Csa5, two possibilities present themselves: selective domain loss from an ancestral Cse2 to create two new subfamilies or domain fusion of two separate families to create a new Cse2 family. A definitive answer awaits structural studies of further small subunits from other CRISPR effector complexes.
Organizational Affiliation:
Biomedical Sciences Research Complex, University of St. Andrews, North Haugh, St. Andrews, Fife, UK.