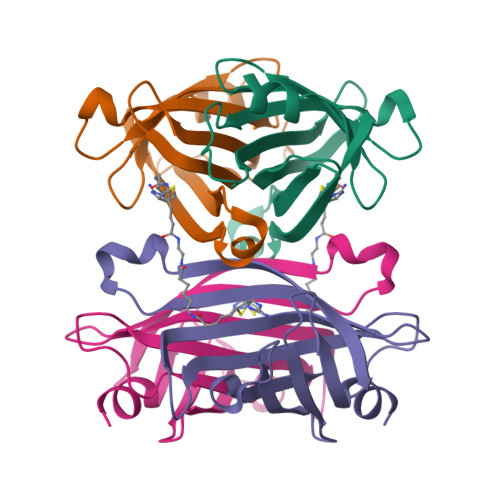
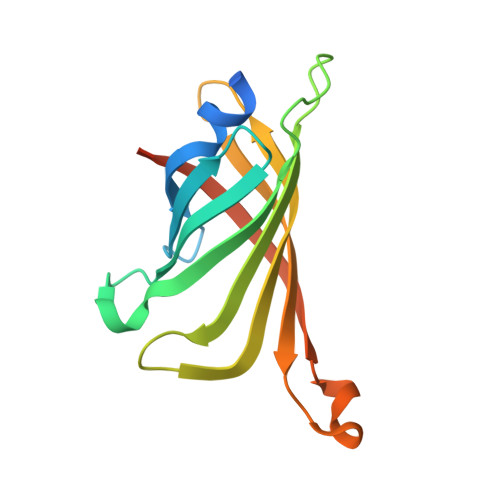
Structure-based design and synthesis of a bivalent iminobiotin analog showing strong affinity toward a low immunogenic streptavidin mutant.
Kawato, T., Mizohata, E., Shimizu, Y., Meshizuka, T., Yamamoto, T., Takasu, N., Matsuoka, M., Matsumura, H., Kodama, T., Kanai, M., Doi, H., Inoue, T., Sugiyama, A.(2015) Biosci Biotechnol Biochem 79: 640-642
- PubMed: 25560769
- DOI: https://doi.org/10.1080/09168451.2014.991692
- Primary Citation of Related Structures:
3X00 - PubMed Abstract:
The streptavidin/biotin interaction has been widely used as a useful tool in research fields. For application to a pre-targeting system, we previously developed a streptavidin mutant that binds to an iminobiotin analog while abolishing affinity for natural biocytin. Here, we design a bivalent iminobiotin analog that shows 1000-fold higher affinity than before, and determine its crystal structure complexed with the mutant protein.
Organizational Affiliation:
a Division of Applied Chemistry, Graduate School of Engineering , Osaka University , Suita , Japan.