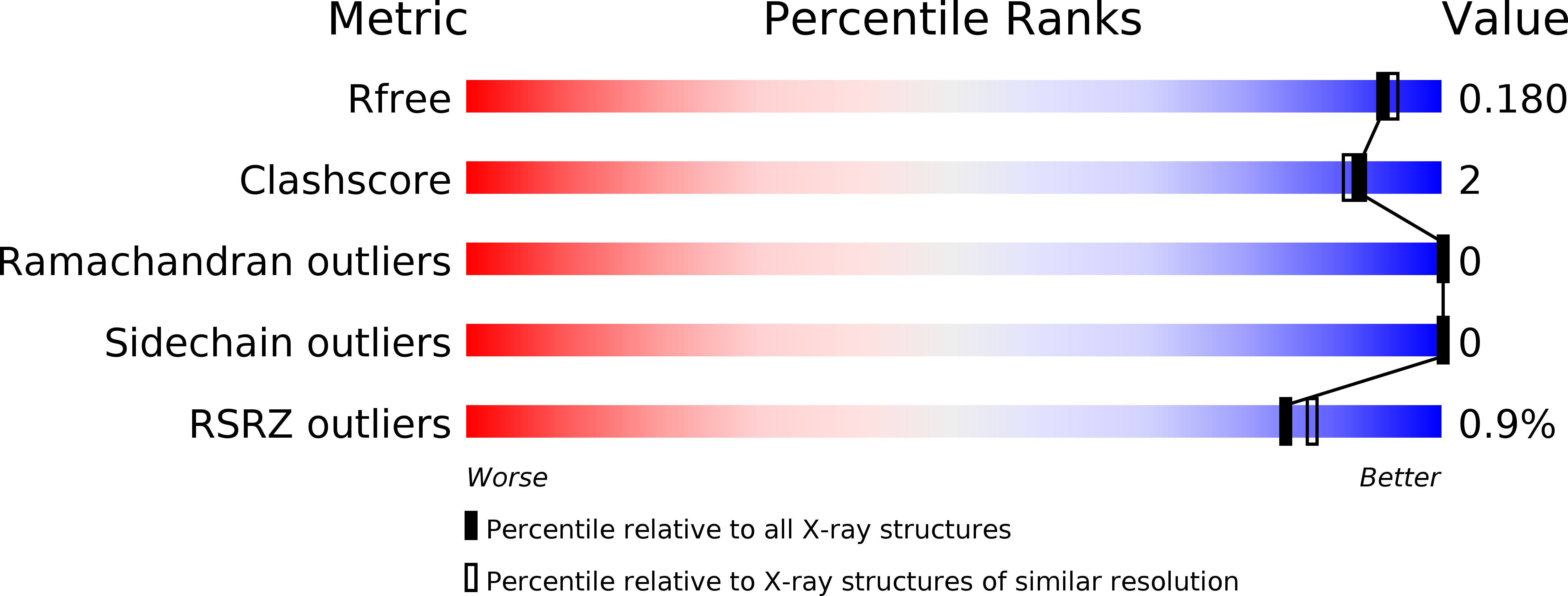
Structural characterization reveals the keratinolytic activity of an arthrobacter nicotinovorans protease.
Sone, T., Haraguchi, Y., Kuwahara, A., Ose, T., Takano, M., Abe, A., Tanaka, M., Tanaka, I., Asano, K.(2015) Protein Pept Lett 22: 63-72
- PubMed: 25256266
- DOI: https://doi.org/10.2174/0929866521666140919100851
- Primary Citation of Related Structures:
3WY8 - PubMed Abstract:
Elevated cadmium (Cd) concentrations in fishery byproducts are an environmental concern, that might be reduced by enzymatic removal and adsorption of the contaminants during recycling the byproducts as animal food. We cloned the gene for Arthrobacter nicotinovorans serine protease (ANISEP), which was isolated from the hepatopancreas of the Japanese scallop (Patiopecten yessoensis) and has been found to be an effective enzyme for Cd(II) removal. The gene is 993 bp in length and encodes 330 amino acids, including the pre (1-30) and pro (31-111) sequences. The catalytic triad consists of His, Asp, and Ser. Sequence similarities indicate that ANISEP is a extracellular serine protease. X-ray crystallography revealed structural similarities between ANISEP and the trypsin-like serine protease NAALP from Nesterenkonia sp. Site-directed mutagenesis identified Ser171 as catalytic residue. The keratinolytic activity of ANISEP was 10-fold greater than that of trypsin. ANISEP digested Cd(II)-bound recombinant metallothionein MT-10a from Laternula elliptica, but did not release Cd. These results further suggest ANISEP is a trypsin-like serine protease that can release Cd from the Japanese scallop hepatopancreas because of its strong keratinolytic activity.
Organizational Affiliation:
Research Faculty of Agriculture, Hokkaido University, Kita-9 Nishi-9, Kita-ku, Sapporo 060-8589, Japan. sonet@chem.agr.hokudai.ac.jp.