The Structural Differences between a Glycoprotein Specific F-Box Protein Fbs1 and Its Homologous Protein FBG3
Kumanomidou, T., Nishio, K., Takagi, K., Nakagawa, T., Suzuki, A., Yamane, T., Tokunaga, F., Iwai, K., Murakami, A., Yoshida, Y., Tanaka, K., Mizushima, T.(2015) PLoS One 10: e0140366-e0140366
- PubMed: 26460611
- DOI: https://doi.org/10.1371/journal.pone.0140366
- Primary Citation of Related Structures:
3WSO - PubMed Abstract:
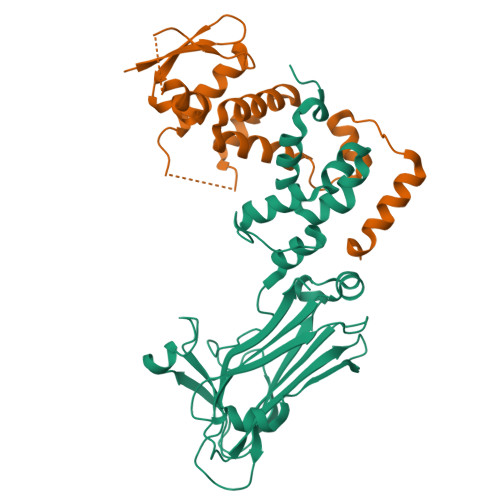
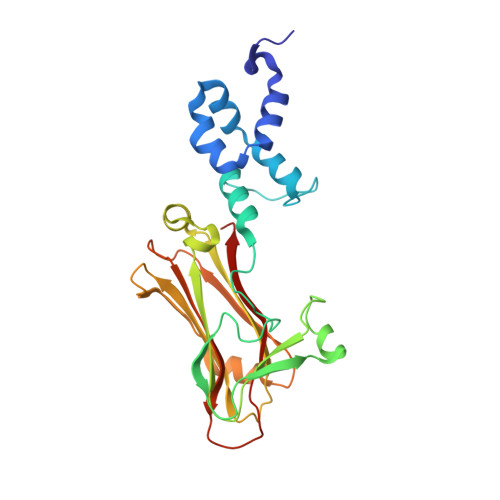
The Skp1-Cul1-F-box protein (SCF) complex catalyzes protein ubiquitination in diverse cellular processes and is one of the best-characterized ubiquitin ligases. F-box proteins determine the substrate specificities of SCF ubiquitin ligases. Among these, Fbs1/FBG1/FBXO2, Fbs2/FBG2/FBXO6, and Fbs3/FBG5/FBXO27 recognize the N-glycans of glycoproteins, whereas FBG3/FBXO44 has no sugar-binding activity, despite the high sequence homology and conservation of the residues necessary for oligosaccharide binding between Fbs1-3 and FBG3. Here we determined the crystal structure of the Skp1-FBG3 complex at a resolution of 2.6 Å. The substrate-binding domain of FBG3 is composed of a 10-stranded antiparallel β-sandwich with three helices. Although the overall structure of FBG3 is similar to that of Fbs1, the residues that form the Fbs1 carbohydrate-binding pocket failed to be superposed with the corresponding residues of FBG3. Structure-based mutational analysis shows that distinct hydrogen bond networks of four FBG3 loops, i.e., β2-β3, β5-β6, β7-β8, and β9-β10, prevent the formation of the carbohydrate-binding pocket shown in Fbs1.
Organizational Affiliation:
Department of Biotechnology, Graduate School of Engineering, Nagoya University, Chikusa-ku, Nagoya, Japan.