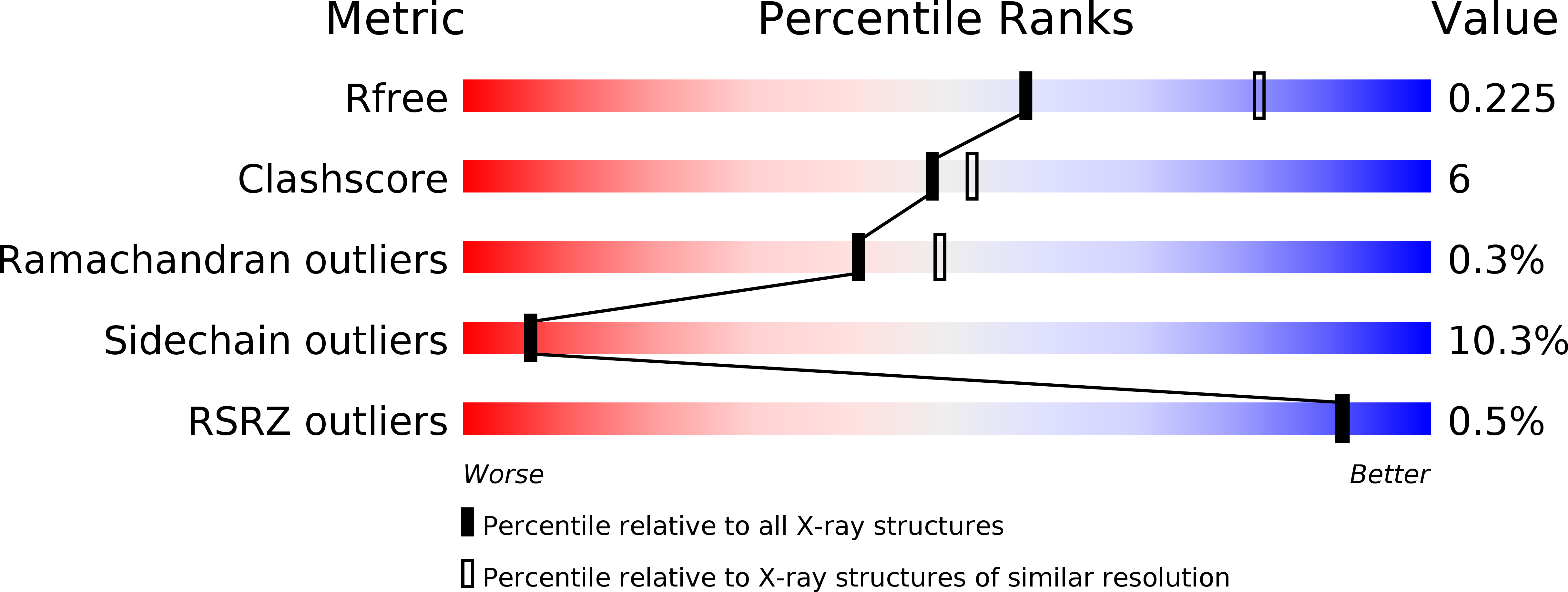
Identification of the HcgB enzyme in [Fe]-hydrogenase-cofactor biosynthesis.
Fujishiro, T., Tamura, H., Schick, M., Kahnt, J., Xie, X., Ermler, U., Shima, S.(2013) Angew Chem Int Ed Engl 52: 12555-12558
- PubMed: 24249552
- DOI: https://doi.org/10.1002/anie.201306745
- Primary Citation of Related Structures:
3WB0, 3WB1, 3WB2
Organizational Affiliation:
Max-Planck-Institut für terrestrische Mikrobiologie, Karl-von-Frisch-Strasse 10, 35043 Marburg (Germany) http://www.mpi-marburg.mpg.de/