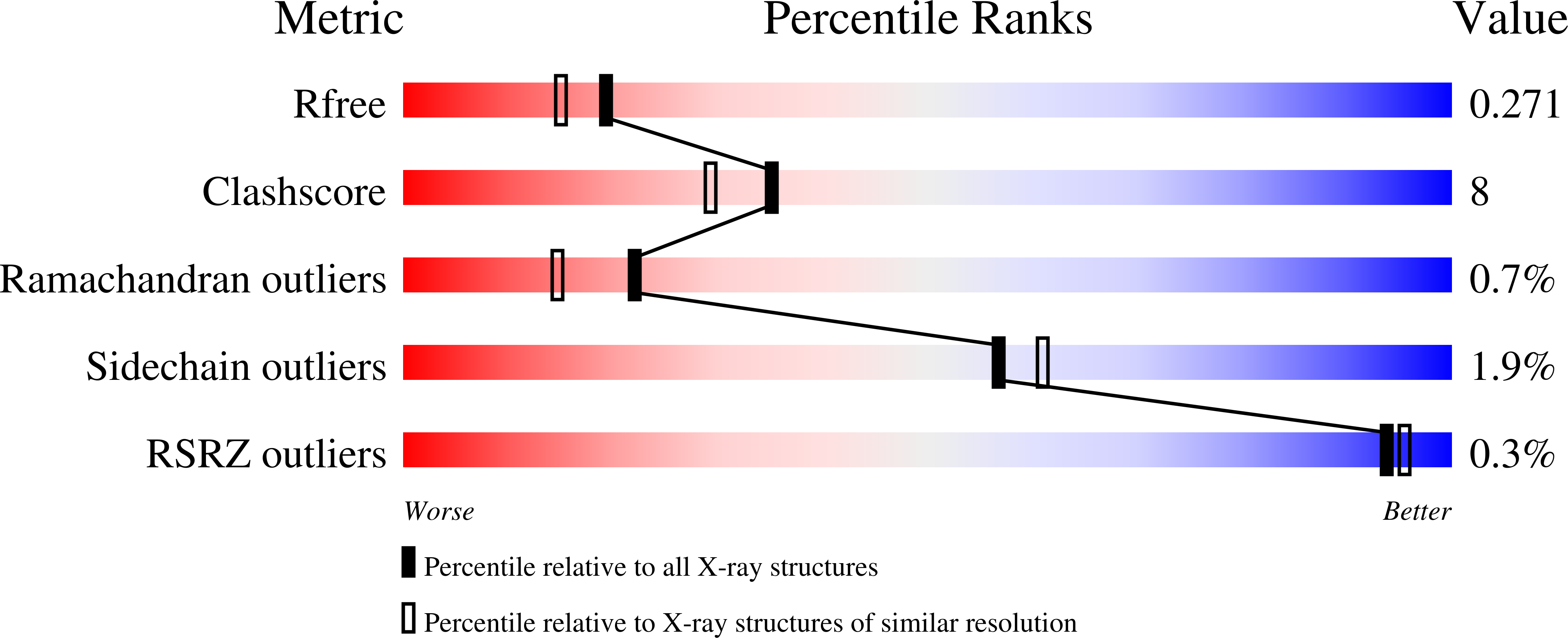
Structural basis for inhibition of carboxypeptidase B by selenium-containing inhibitor: selenium coordinates to zinc in enzyme.
Yoshimoto, N., Itoh, T., Inaba, Y., Ishii, H., Yamamoto, K.(2013) J Med Chem 56: 7527-7535
- PubMed: 24010887
- DOI: https://doi.org/10.1021/jm400816v
- Primary Citation of Related Structures:
3WAB, 3WC5, 3WC6, 3WC7 - PubMed Abstract:
Activated thrombin-activatable fibrinolysis inhibitor (TAFIa) is a zinc-containing carboxypeptidase and significantly inhibits fibrinolysis. TAFIa inhibitors are thus expected to act as profibrinolytic agents. We recently reported the design and synthesis of selenium-containing inhibitors of TAFIa and their inhibitory activity. Here we report the crystal structures of potent selenium-, sulfur-, and phosphinic acid-containing inhibitors bound to porcine pancreatic carboxypeptidase B (ppCPB). ppCPB is a TAFIa homologue and is surrogate TAFIa for crystallographic analysis. Crystal structures of ppCPB complexed with selenium compound 1a, its sulfur analogue 2, and phosphinic acid derivative EF6265 were determined at 1.70, 2.15, and 1.90 Å resolution, respectively. Each inhibitor binds to the active site of ppCPB in a similar manner to that observed for previously reported inhibitors. Thus, in complexes, selenium, sulfur, and phosphinic acid oxygen coordinate to zinc in ppCPB. This is the first observation and report of selenium coordinating to zinc in CPB.
Organizational Affiliation:
High Technology Research Center, ‡Laboratory of Drug Design and Medicinal Chemistry and §Laboratory of Molecular and Cellular Pathophysiology, Showa Pharmaceutical University , 3-3165 Higashi-Tamagawagakuen, Machida, Tokyo 194-8543, Japan.