Effects of argnine residue around the substrate pocket on the substrate specificity of thiocyanate hydrolase
Yamanaka, Y., Sato, M., Arakawa, T., Namima, S., Hori, S., Ohtaki, A., Noguchi, K., Katayama, Y., Yohda, M., Odaka, M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Thiocyanate hydrolase subunit alpha | 126 | Thiobacillus thioparus | Mutation(s): 0 Gene Names: scnA EC: 3.5.5.8 | 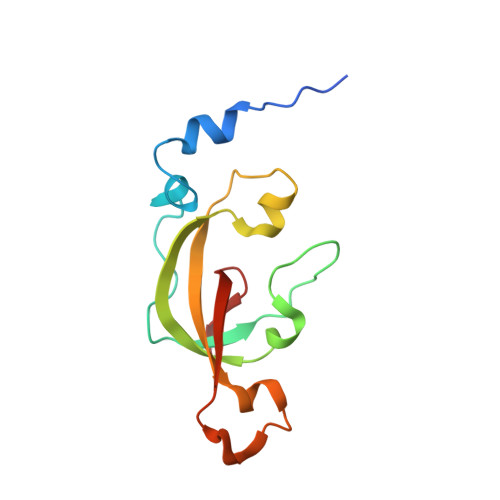 | |
UniProt | |||||
Find proteins for O66187 (Thiobacillus thioparus) Explore O66187 Go to UniProtKB: O66187 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O66187 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Thiocyanate hydrolase subunit beta | 157 | Thiobacillus thioparus | Mutation(s): 0 Gene Names: scnB EC: 3.5.5.8 | 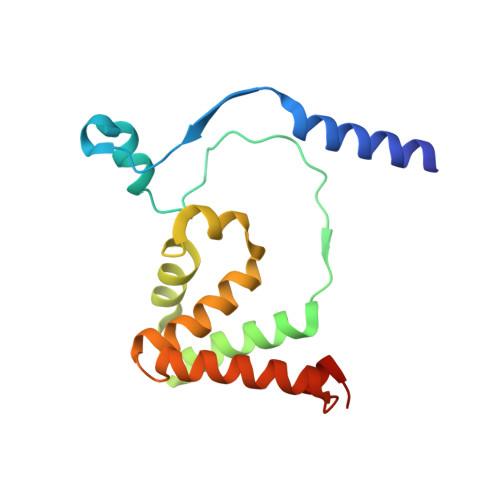 | |
UniProt | |||||
Find proteins for O66186 (Thiobacillus thioparus) Explore O66186 Go to UniProtKB: O66186 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O66186 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Thiocyanate hydrolase subunit gamma | 243 | Thiobacillus thioparus | Mutation(s): 1 Gene Names: scnC EC: 3.5.5.8 | 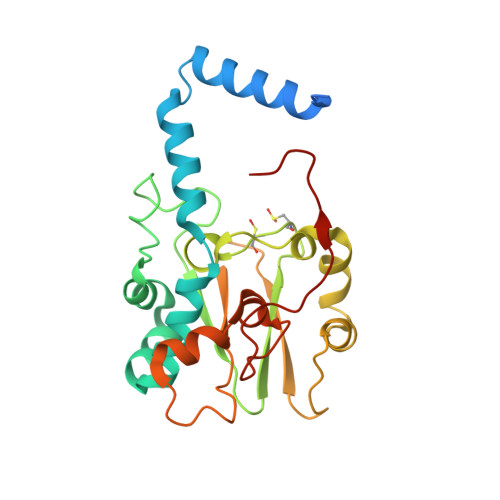 | |
UniProt | |||||
Find proteins for O66188 (Thiobacillus thioparus) Explore O66188 Go to UniProtKB: O66188 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O66188 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| TLA Query on TLA | N [auth C], P [auth F], R [auth I], T [auth L] | L(+)-TARTARIC ACID C4 H6 O6 FEWJPZIEWOKRBE-JCYAYHJZSA-N |  | ||
| 3CO Query on 3CO | M [auth C], O [auth F], Q [auth I], S [auth L] | COBALT (III) ION Co JAWGVVJVYSANRY-UHFFFAOYSA-N |  | ||
| Modified Residues 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSD Query on CSD | C, F, I, L | L-PEPTIDE LINKING | C3 H7 N O4 S |  | CYS |
| CSO Query on CSO | C, F, I, L | L-PEPTIDE LINKING | C3 H7 N O3 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 113.448 | α = 90 |
| b = 172.398 | β = 90 |
| c = 172.531 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |