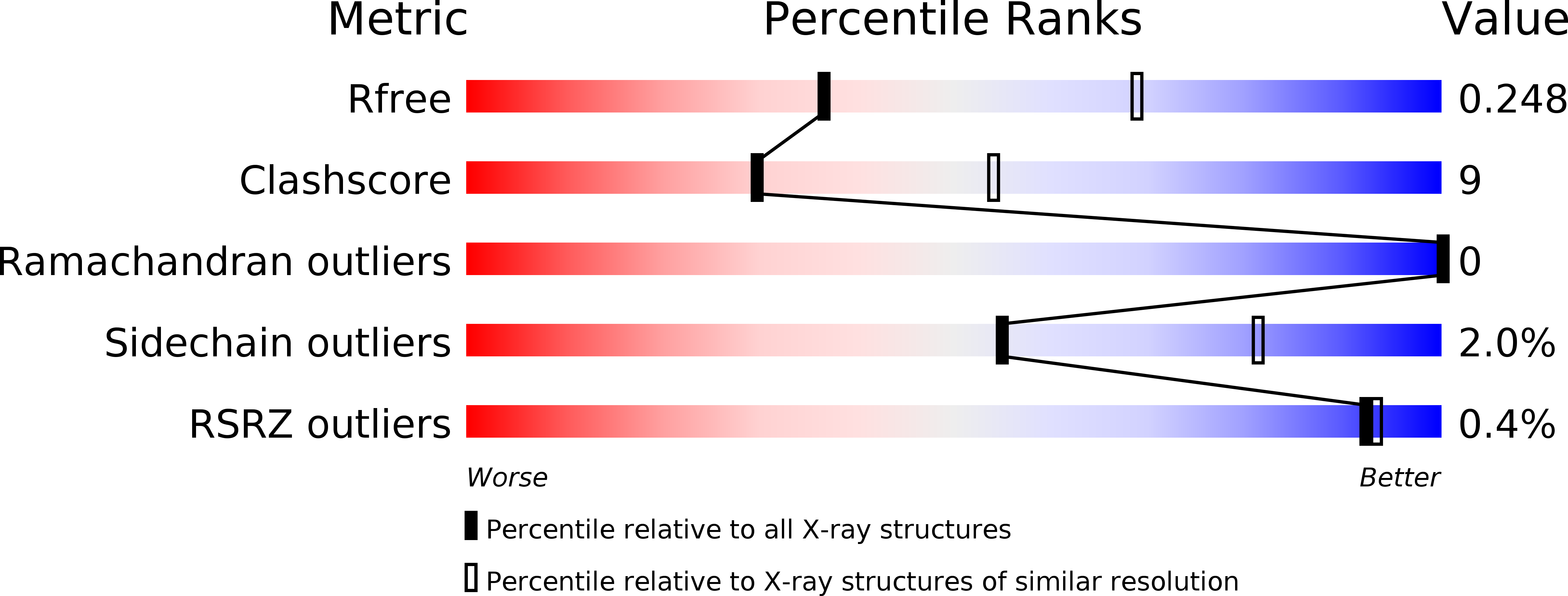
Boundary of the Nucleotide-Binding Domain of Streptococcus ComA Based on Functional and Structural Analysis
Ishii, S., Yano, T., Okamoto, A., Murakawa, T., Hayashi, H.(2013) Biochemistry 52: 2545-2555
- PubMed: 23534432
- DOI: https://doi.org/10.1021/bi3017069
- Primary Citation of Related Structures:
3VX4 - PubMed Abstract:
The ATP-binding cassette (ABC) transporter ComA is a key molecule essential for the first step of the quorum-sensing system of Streptococcus. The nucleotide binding domains (NBD) of Streptococcus mutans ComA with different N termini, NBD1 (amino acid residues 495-760), NBD2 (517-760), and NBD3 (528-760), were expressed, purified, and characterized. The shortest NBD3 corresponds to the region commonly defined as NBD in the database searches of ABC transporters. A kinetic analysis showed that the extra N-terminal region conferred a significantly higher ATP hydrolytic activity on the NBD at a neutral pH. Gel-filtration, X-ray crystallography, and mutational analyses suggest that at least four to five residues beyond the N-terminal boundary of NBD3 indeed participate in stabilizing the protein scaffold of the domain structure, thereby facilitating the ATP-dependent dimerization of NBD which is a prerequisite to the catalysis. These findings, together with the presence of a highly conserved glycine residue in this region, support the redefinition of the N-terminal boundary of the NBD of these types of ABC exporters.
Organizational Affiliation:
Department of Biochemistry, Faculty of Nursing, Osaka Medical College, Osaka 569-8686, Japan. med002@art.osaka-med.ac.jp