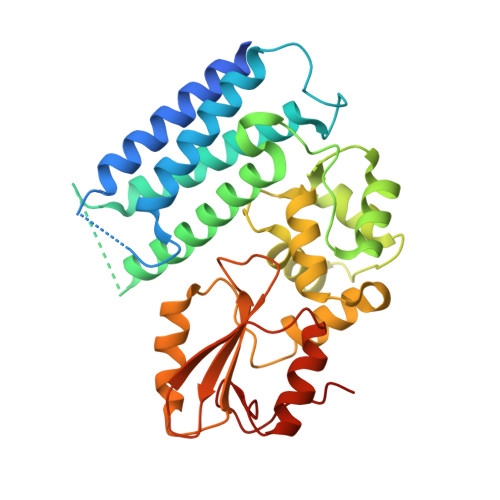
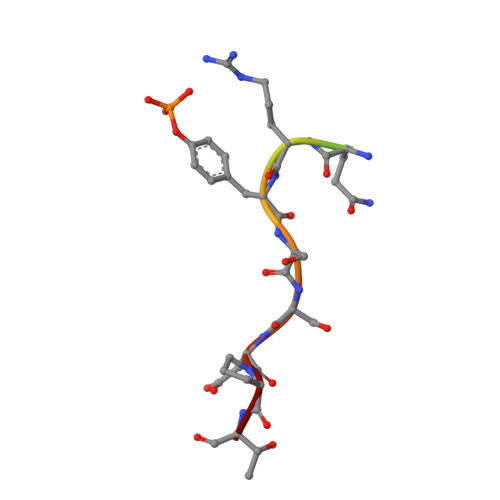
Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
Takeshita, K., Tezuka, T., Isozaki, Y., Yamashita, E., Suzuki, M., Kim, M., Yamanashi, Y., Yamamoto, T., Nakagawa, A.(2012) J Biochem 152: 487-495
- PubMed: 22888118
- DOI: https://doi.org/10.1093/jb/mvs085
- Primary Citation of Related Structures:
3VRN, 3VRO, 3VRP, 3VRQ, 3VRR - PubMed Abstract:
Through their ubiquitin ligase activity, Cbl-family proteins suppress signalling mediated by protein-tyrosine kinases (PTKs), but can also function as adaptor proteins to positively regulate signalling. The tyrosine kinase binding (TKB) domain of this family is critical for binding with tyrosine-phosphorylated target proteins. Here, we analysed the crystal structure of the TKB domain of Cbl-c/Cbl-3 (Cbl-c TKB), which is a distinct member of the mammalian Cbl-family. In comparison with Cbl TKB, Cbl-c TKB showed restricted structural flexibility upon phosphopeptide binding. A mutation in Cbl-c TKB augmenting this flexibility enhanced its binding to target phosphoproteins. These results suggest that proteins, post-translational modifications or mutations that alter structural flexibility of the TKB domain of Cbl-family proteins could regulate their binding to target phosphoproteins and thereby, affect PTK-mediated signalling.
Organizational Affiliation:
Institute for Protein Research, Osaka University, Suita, Osaka 565-0871, Japan.