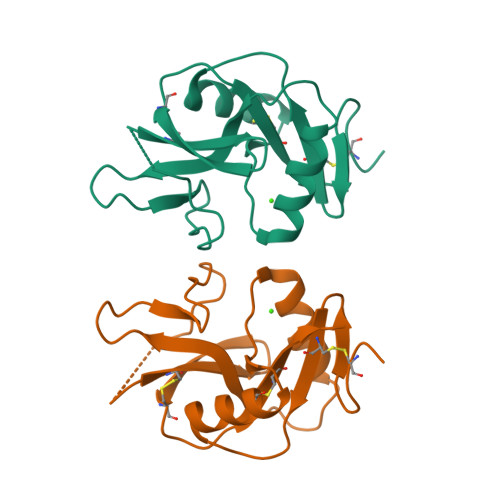
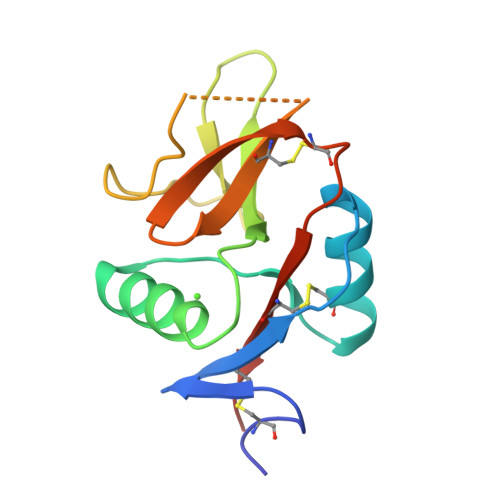
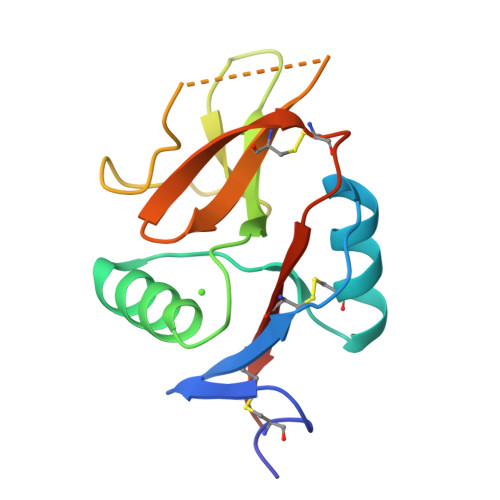
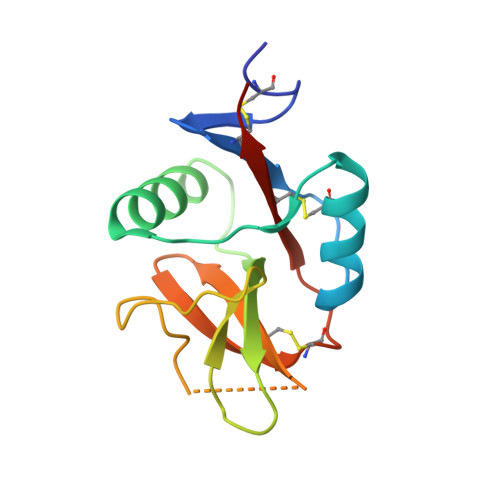
The dendritic cell receptor Clec9A binds damaged cells via exposed actin filaments.
Zhang, J.G., Czabotar, P.E., Policheni, A.N., Caminschi, I., Wan, S.S., Kitsoulis, S., Tullett, K.M., Robin, A.Y., Brammananth, R., van Delft, M.F., Lu, J., O'Reilly, L.A., Josefsson, E.C., Kile, B.T., Chin, W.J., Mintern, J.D., Olshina, M.A., Wong, W., Baum, J., Wright, M.D., Huang, D.C., Mohandas, N., Coppel, R.L., Colman, P.M., Nicola, N.A., Shortman, K., Lahoud, M.H.(2012) Immunity 36: 646-657
- PubMed: 22483802
- DOI: https://doi.org/10.1016/j.immuni.2012.03.009
- Primary Citation of Related Structures:
3VPP - PubMed Abstract:
The immune system must distinguish viable cells from cells damaged by physical and infective processes. The damaged cell-recognition molecule Clec9A is expressed on the surface of the mouse and human dendritic cell subsets specialized for the uptake and processing of material from dead cells. Clec9A recognizes a conserved component within nucleated and nonnucleated cells, exposed when cell membranes are damaged. We have identified this Clec9A ligand as a filamentous form of actin in association with particular actin-binding domains of cytoskeletal proteins. We have determined the crystal structure of the human CLEC9A C-type lectin domain and propose a functional dimeric structure with conserved tryptophans in the ligand recognition site. Mutation of these residues ablated CLEC9A binding to damaged cells and to the isolated ligand complexes. We propose that Clec9A provides targeted recruitment of the adaptive immune system during infection and can also be utilized to enhance immune responses generated by vaccines.
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria 3052, Australia.