Structural and enzymatic characterization of BacD, an l-amino acid dipeptide ligase from Bacillus subtilis
Shomura, Y., Hinokuchi, E., Ikeda, H., Senoo, A., Takahashi, Y., Saito, J., Komori, H., Shibata, N., Yonetani, Y., Higuchi, Y.(2012) Protein Sci
- PubMed: 22407814
- DOI: https://doi.org/10.1002/pro.2058
- Primary Citation of Related Structures:
3VMM - PubMed Abstract:
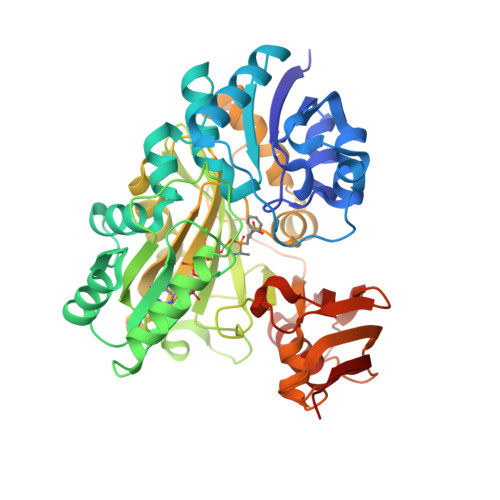
BacD is an ATP-dependent dipeptide ligase responsible for the biosynthesis of L-alanyl-L-anticapsin, a precursor of an antibiotic produced by Bacillus spp. In contrast to the well-studied and phylogenetically related D-alanine: D-alanine ligase (Ddl), BacD synthesizes dipeptides using L-amino acids as substrates and has a low substrate specificity in vitro. The enzyme is of great interest because of its potential application in industrial protein engineering for the environmentally friendly biological production of useful peptide compounds, such as physiologically active peptides, artificial sweeteners and antibiotics, but the determinants of its substrate specificity and its catalytic mechanism have not yet been established due to a lack of structural information. In this study, we report the crystal structure of BacD in complex with ADP and an intermediate analog, phosphorylated phosphinate L-alanyl-L-phenylalanine, refined to 2.5-Å resolution. The complex structure reveals that ADP and two magnesium ions bind in a manner similar to that of Ddl. However, the dipeptide orientation is reversed, and, concomitantly, the entrance to the amino acid binding cavity differs in position. Enzymatic characterization of two mutants, Y265F and S185A, demonstrates that these conserved residues are not catalytic residues at least in the reaction where L-phenylalanine is used as a substrate. On the basis of the biochemical and the structural data, we propose a reaction scheme and a catalytic mechanism for BacD.
Organizational Affiliation:
Department of Life Science, Graduate School of Life Science, University of Hyogo, 3-2-1 Koto, Ako-gun, Hyogo 678-1297, Japan.