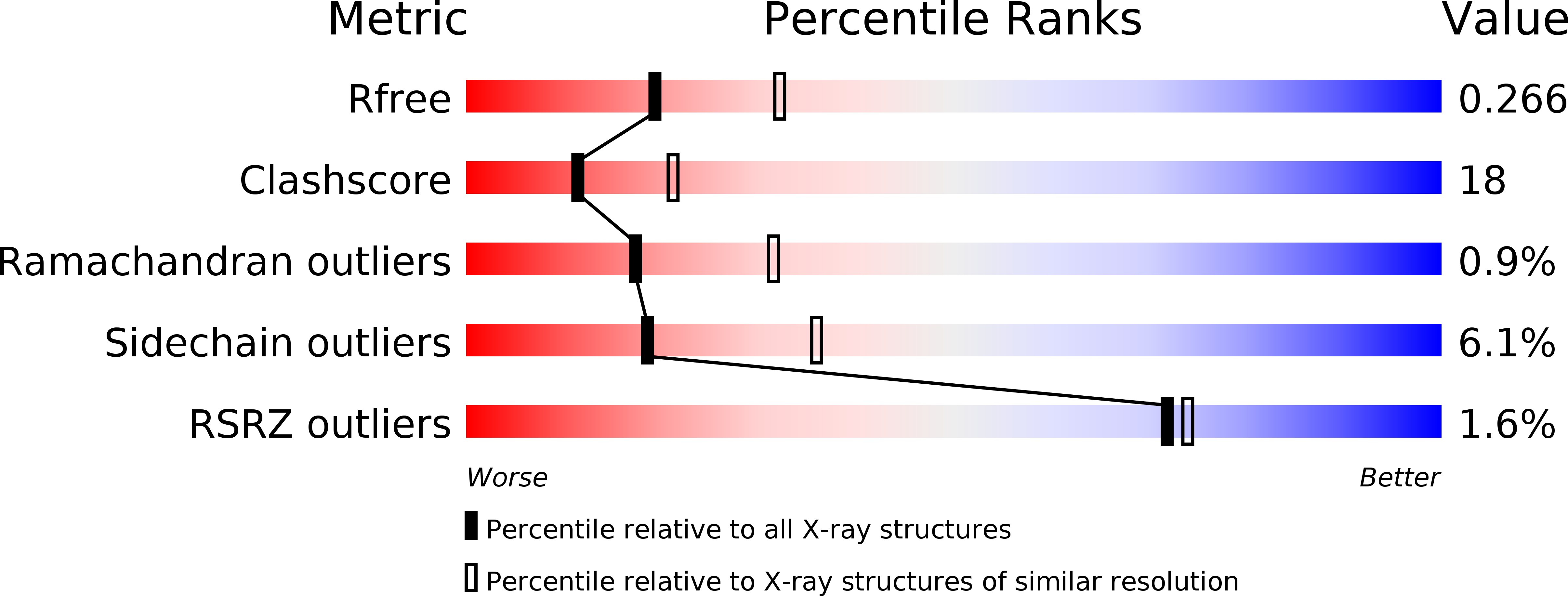
A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Arai, S., Yonezawa, Y., Okazaki, N., Matsumoto, F., Tamada, T., Tokunaga, H., Ishibashi, M., Blaber, M., Tokunaga, M., Kuroki, R.(2012) Protein Sci 21: 498-510
- PubMed: 22275000
- DOI: https://doi.org/10.1002/pro.2032
- Primary Citation of Related Structures:
3VGS, 3VGT, 3VGU, 3VGV - PubMed Abstract:
Nucleoside diphosphate kinase (NDK) is known to form homotetramers or homohexamers. To clarify the oligomer state of NDK from moderately halophilic Halomonas sp. 593 (HaNDK), the oligomeric state of HaNDK was characterized by light scattering followed by X-ray crystallography. The molecular weight of HaNDK is 33,660, and the X-ray crystal structure determination to 2.3 and 2.7 Å resolution showed a dimer form which was confirmed in the different space groups of R3 and C2 with an independent packing arrangement. This is the first structural evidence that HaNDK forms a dimeric assembly. Moreover, the inferred molecular mass of a mutant HaNDK (E134A) indicated 62.1-65.3 kDa, and the oligomerization state was investigated by X-ray crystallography to 2.3 and 2.5 Å resolution with space groups of P2(1) and C2. The assembly form of the E134A mutant HaNDK was identified as a Type I tetramer as found in Myxococcus NDK. The structural comparison between the wild-type and E134A mutant HaNDKs suggests that the change from dimer to tetramer is due to the removal of negative charge repulsion caused by the E134 in the wild-type HaNDK. The higher ordered association of proteins usually contributes to an increase in thermal stability and substrate affinity. The change in the assembly form by a minimum mutation may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
Organizational Affiliation:
Molecular Structural Biology Group, Quantum Beam Science Directorate, Japan Atomic Energy Agency, 2-4 Shirakata-Shirane, Tokai, Ibaraki 319-1195, Japan.