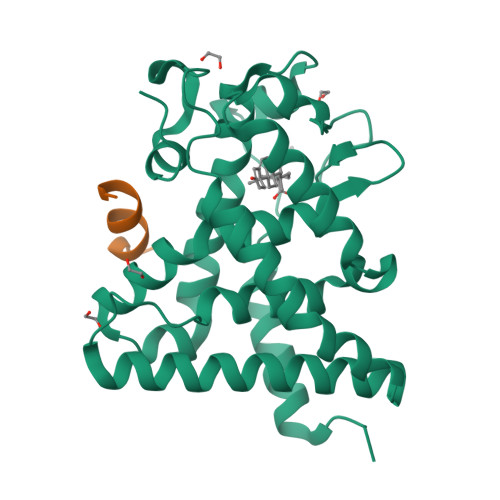
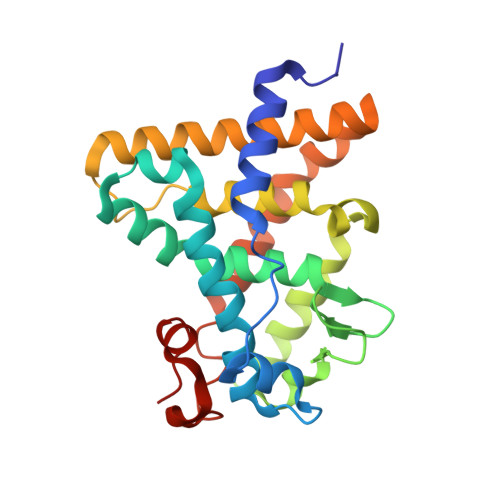
Structural Conservation of Ligand Binding Reveals a Bile Acid-like Signaling Pathway in Nematodes.
Zhi, X., Zhou, X.E., Melcher, K., Motola, D.L., Gelmedin, V., Hawdon, J., Kliewer, S.A., Mangelsdorf, D.J., Xu, H.E.(2012) J Biological Chem 287: 4894-4903
- PubMed: 22170062
- DOI: https://doi.org/10.1074/jbc.M111.315242
- Primary Citation of Related Structures:
3UP0, 3UP3 - PubMed Abstract:
Bile acid-like molecules named dafachronic acids (DAs) control the dauer formation program in Caenorhabditis elegans through the nuclear receptor DAF-12. This mechanism is conserved in parasitic nematodes to regulate their dauer-like infective larval stage, and as such, the DAF-12 ligand binding domain has been identified as an important therapeutic target in human parasitic hookworm species that infect more than 600 million people worldwide. Here, we report two x-ray crystal structures of the hookworm Ancylostoma ceylanicum DAF-12 ligand binding domain in complex with DA and cholestenoic acid (a bile acid-like metabolite), respectively. Structure analysis and functional studies reveal key residues responsible for species-specific ligand responses of DAF-12. Furthermore, DA binds to DAF-12 mechanistically and is structurally similar to bile acids binding to the mammalian bile acid receptor farnesoid X receptor. Activation of DAF-12 by cholestenoic acid and the cholestenoic acid complex structure suggest that bile acid-like signaling pathways have been conserved in nematodes and mammals. Together, these results reveal the molecular mechanism for the interplay between parasite and host, provide a structural framework for DAF-12 as a promising target in treating nematode parasitism, and provide insight into the evolution of gut parasite hormone-signaling pathways.
Organizational Affiliation:
Laboratory of Structural Sciences, Van Andel Research Institute, Grand Rapids, Michigan 49503, USA.