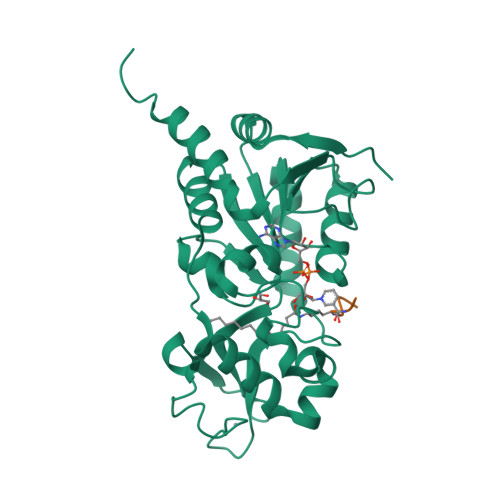
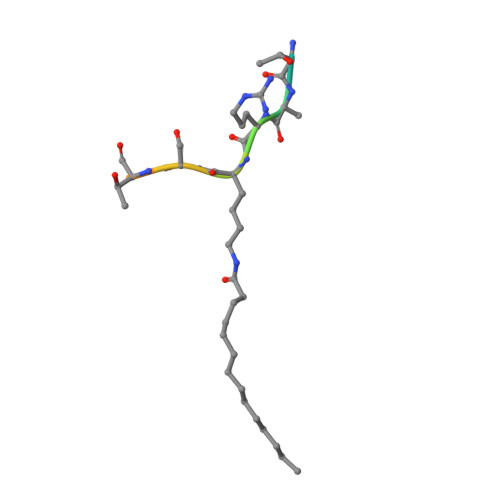
Plasmodium falciparum Sir2A Preferentially Hydrolyzes Medium and Long Chain Fatty Acyl Lysine
Zhu, A.Y., Zhou, Y., Khan, S., Deitsch, K.W., Hao, Q., Lin, H.(2011) ACS Chem Biol
- PubMed: 21992006
- DOI: https://doi.org/10.1021/cb200230x
- Primary Citation of Related Structures:
3U31, 3U3D - PubMed Abstract:
Plasmodium falciparum Sir2A (PfSir2A), a member of the sirtuin family of nicotinamide adenine dinucleotide-dependent deacetylases, has been shown to regulate the expression of surface antigens to evade the detection by host immune surveillance. It is thought that PfSir2A achieves this by deacetylating histones. However, the deacetylase activity of PfSir2A is weak. Here we present enzymology and structural evidence supporting that PfSir2A catalyzes the hydrolysis of medium and long chain fatty acyl groups from lysine residues more efficiently. Furthermore, P. falciparum proteins are found to contain such fatty acyl lysine modifications that can be removed by purified PfSir2A in vitro. Together, the data suggest that the physiological function of PfSir2A in antigen variation may be achieved by removing medium and long chain fatty acyl groups from protein lysine residues. The robust activity of PfSir2A would also facilitate the development of PfSir2A inhibitors, which may have therapeutic value in malaria treatment.