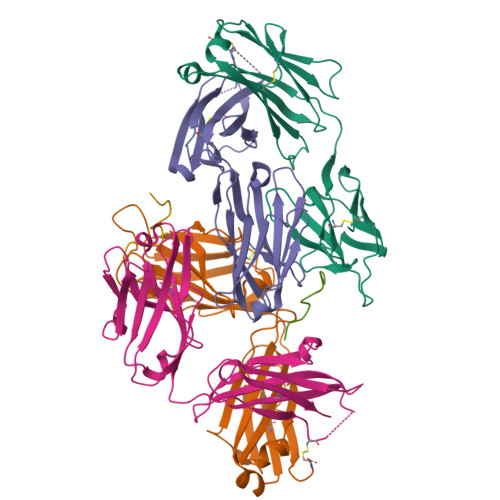
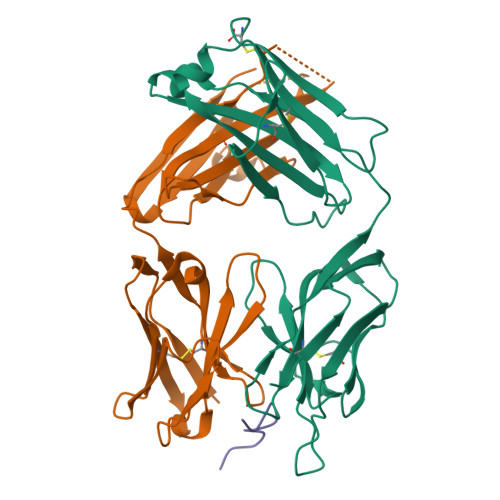
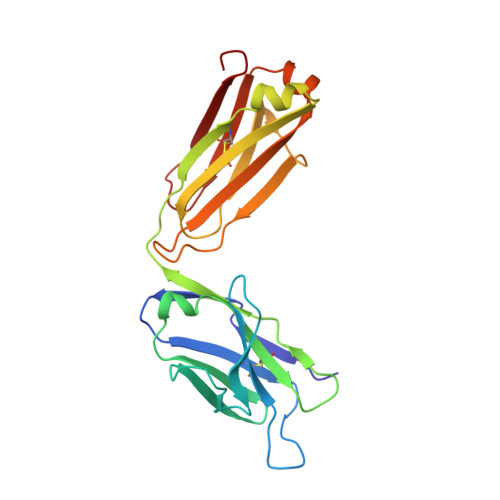
Structural Basis of C-terminal beta-Amyloid Peptide Binding by the Antibody Ponezumab for the Treatment of Alzheimer's Disease
La Porte, S.L., Bollini, S.S., Lanz, T.A., Abdiche, Y.N., Rusnak, A.S., Ho, W.H., Kobayashi, D., Harrabi, O., Pappas, D., Mina, E.W., Milici, A.J., Kawabe, T.T., Bales, K., Lin, J.C., Pons, J.(2012) J Mol Biology 421: 525-536
- PubMed: 22197375
- DOI: https://doi.org/10.1016/j.jmb.2011.11.047
- Primary Citation of Related Structures:
3U0T - PubMed Abstract:
Alzheimer's disease, the most common cause of dementia in the elderly and characterized by the deposition and accumulation of plaques, is composed in part of β-amyloid (Aβ) peptides, loss of neurons, and the accumulation of neurofibrillary tangles. Here, we describe ponezumab, a humanized monoclonal antibody, and show how it binds specifically to the carboxyl (C)-terminus of Aβ40. Ponezumab can label Aβ that is deposited in brain parenchyma found in sections from Alzheimer's disease casualties and in transgenic mouse models that overexpress Aβ. Importantly, ponezumab does not label full-length, non-cleaved amyloid precursor protein on the cell surface. The C-terminal epitope of the soluble Aβ present in the circulation appears to be available for ponezumab binding because systemic administration of ponezumab greatly elevates plasma Aβ40 levels in a dose-dependent fashion after administration to a mouse model that overexpress human Aβ. Administration of ponezumab to transgenic mice also led to a dose-dependent reduction in hippocampal amyloid load. To further explore the nature of ponezumab binding to Aβ40, we determined the X-ray crystal structure of ponezumab in complex with Aβ40 and found that the Aβ40 carboxyl moiety makes extensive contacts with ponezumab. Furthermore, the structure-function analysis supported this critical requirement for carboxy group of AβV40 in the Aβ-ponezumab interaction. These findings provide novel structural insights into the in vivo conformation of the C-terminus of Aβ40 and the brain Aβ-lowering efficacy that we observed following administration of ponezumab in transgenic mouse models.
Organizational Affiliation:
Rinat, Pfizer Inc., South San Francisco, CA 94080, USA.