2,6-Naphthyridines as potent and selective inhibitors of the novel protein kinase C isozymes.
van Eis, M.J., Evenou, J.P., Floersheim, P., Gaul, C., Cowan-Jacob, S.W., Monovich, L., Rummel, G., Schuler, W., Stark, W., Strauss, A., Matt, A., Vangrevelinghe, E., Wagner, J., Soldermann, N.(2011) Bioorg Med Chem Lett 21: 7367-7372
- PubMed: 22078216
- DOI: https://doi.org/10.1016/j.bmcl.2011.10.025
- Primary Citation of Related Structures:
3TXO - PubMed Abstract:
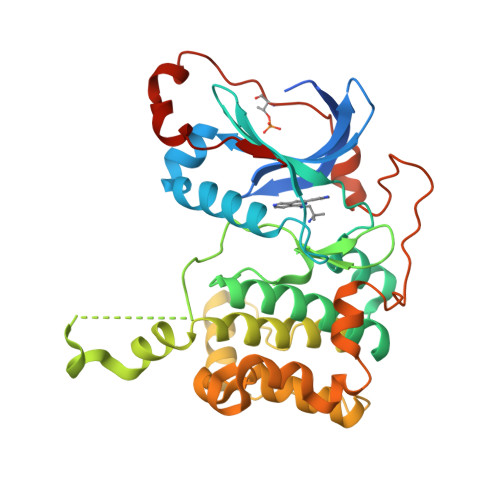
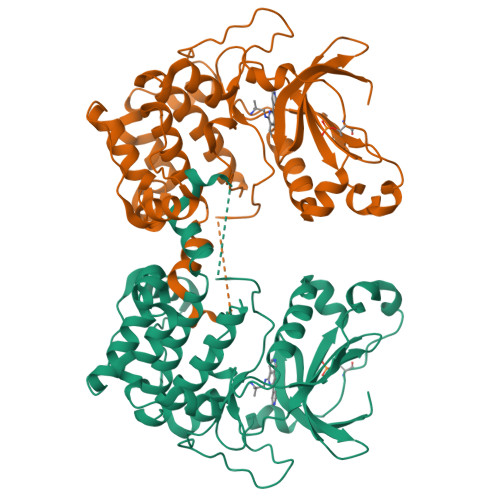
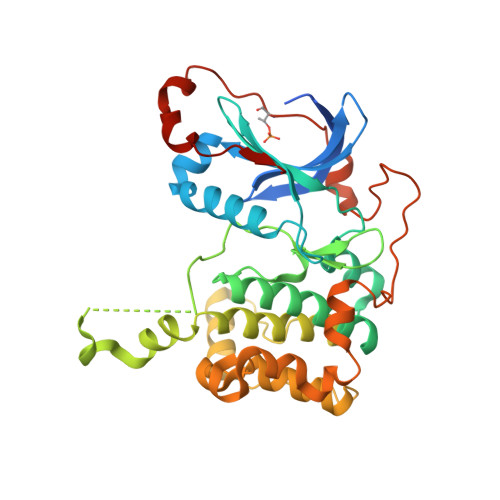
The present study describes a novel series of ATP-competitive PKC inhibitors based on the 2,6-naphthyridine template. Example compounds potently inhibit the novel Protein Kinase C (PKC) isotypes δ, ε, η, θ (in particular PKCε/η, and display a 10-100-fold selectivity over the classical PKC isotypes. The prototype compound 11 was found to inhibit PKCθ-dependent pathways in vitro and in vivo. In vitro, a-CD3/a-CD28-induced lymphocyte proliferation could be effectively blocked in 10% rat whole blood. In mice, 11 dose-dependently inhibited Staphylococcus aureus enterotoxin B-triggered IL-2 serum levels after oral dosing.
Organizational Affiliation:
Novartis Institutes for BioMedical Research, Forum 1, Novartis Campus, CH-4056 Basel, Switzerland. maurice.van_eis@novartis.com