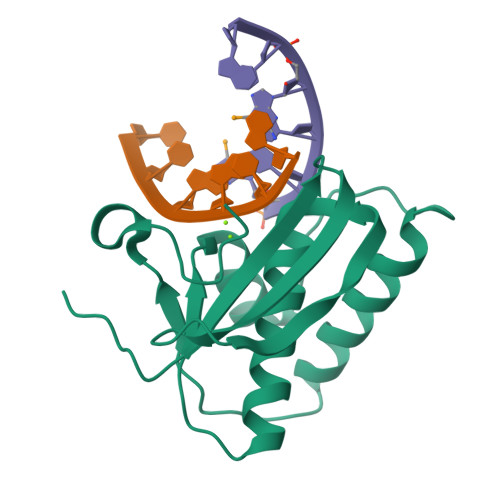
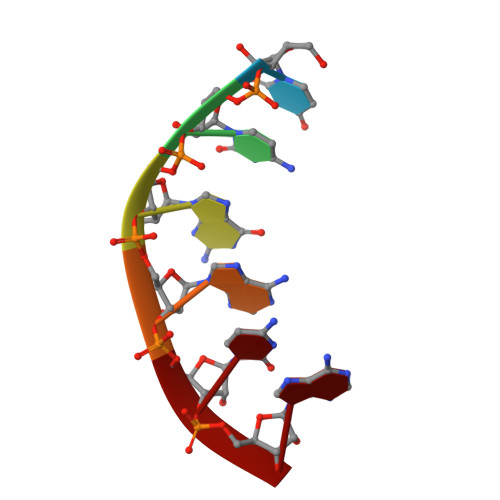
Novel complex MAD phasing and RNase H structural insights using selenium oligonucleotides.
Abdur, R., Gerlits, O.O., Gan, J., Jiang, J., Salon, J., Kovalevsky, A.Y., Chumanevich, A.A., Weber, I.T., Huang, Z.(2014) Acta Crystallogr D Biol Crystallogr 70: 354-361
- PubMed: 24531469
- DOI: https://doi.org/10.1107/S1399004713027922
- Primary Citation of Related Structures:
3TWH - PubMed Abstract:
The crystal structures of protein-nucleic acid complexes are commonly determined using selenium-derivatized proteins via MAD or SAD phasing. Here, the first protein-nucleic acid complex structure determined using selenium-derivatized nucleic acids is reported. The RNase H-RNA/DNA complex is used as an example to demonstrate the proof of principle. The high-resolution crystal structure indicates that this selenium replacement results in a local subtle unwinding of the RNA/DNA substrate duplex, thereby shifting the RNA scissile phosphate closer to the transition state of the enzyme-catalyzed reaction. It was also observed that the scissile phosphate forms a hydrogen bond to the water nucleophile and helps to position the water molecule in the structure. Consistently, it was discovered that the substitution of a single O atom by a Se atom in a guide DNA sequence can largely accelerate RNase H catalysis. These structural and catalytic studies shed new light on the guide-dependent RNA cleavage.
Organizational Affiliation:
Department of Chemistry and Department of Biology, Georgia State University, Atlanta, GA 30303, USA.