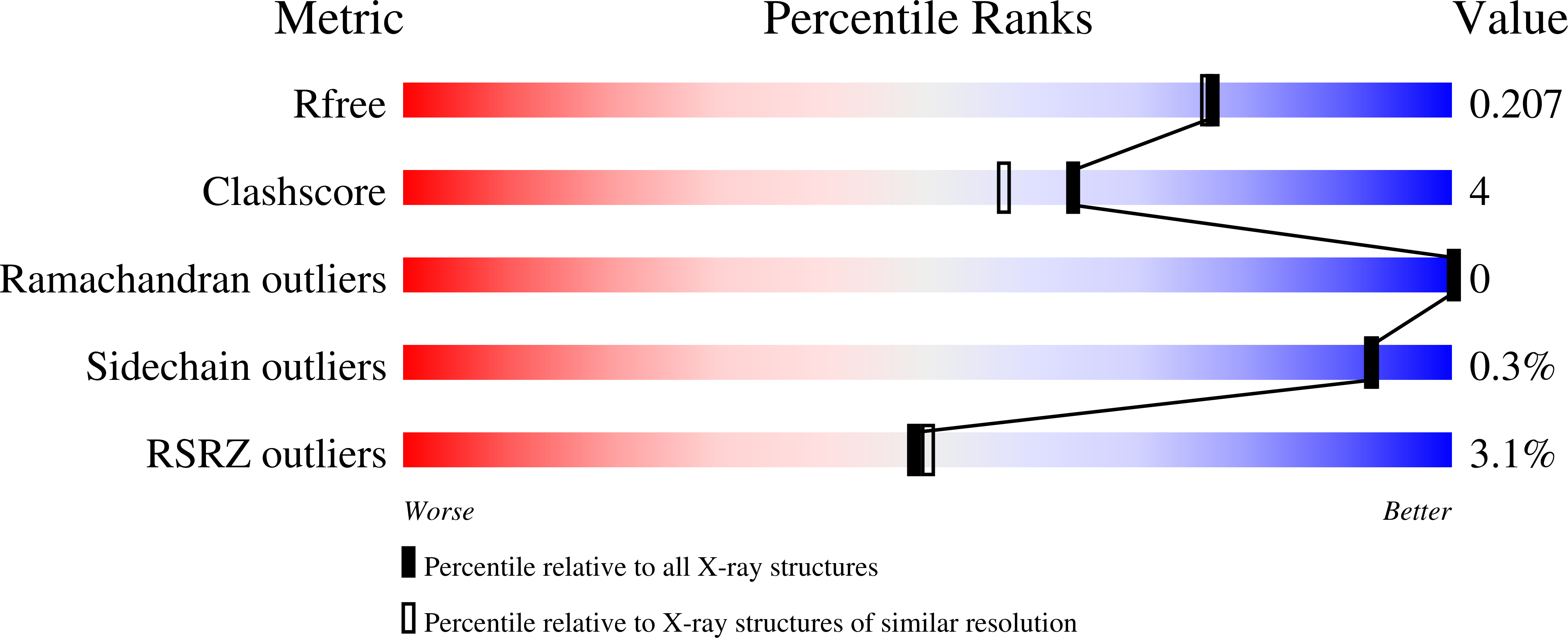
High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
Sanchez, E.J., Lewis, K.M., Danna, B.R., Kang, C.(2012) J Biol Chem 287: 11592-11601
- PubMed: 22337878
- DOI: https://doi.org/10.1074/jbc.M111.335075
- Primary Citation of Related Structures:
3TRP, 3UOM - PubMed Abstract:
Calsequestrin, the major calcium storage protein in both cardiac and skeletal muscle, binds large amounts of Ca(2+) in the sarcoplasmic reticulum and releases them during muscle contraction. For the first time, the crystal structures of Ca(2+) complexes for both human (hCASQ1) and rabbit (rCASQ1) skeletal calsequestrin were determined, clearly defining their Ca(2+) sequestration capabilities through resolution of high- and low-affinity Ca(2+)-binding sites. rCASQ1 crystallized in low CaCl(2) buffer reveals three high-affinity Ca(2+) sites with trigonal bipyramidal, octahedral, and pentagonal bipyramidal coordination geometries, along with three low-affinity Ca(2+) sites. hCASQ1 crystallized in high CaCl(2) shows 15 Ca(2+) ions, including the six Ca(2+) ions in rCASQ1. Most of the low-affinity sites, some of which are μ-carboxylate-bridged, are established by the rotation of dimer interfaces, indicating cooperative Ca(2+) binding that is consistent with our atomic absorption spectroscopic data. On the basis of these findings, we propose a mechanism for the observed in vitro and in vivo dynamic high-capacity and low-affinity Ca(2+)-binding activity of calsequestrin.
Organizational Affiliation:
School of Molecular Biosciences, Washington State University, Pullman, Washington 99164, USA.