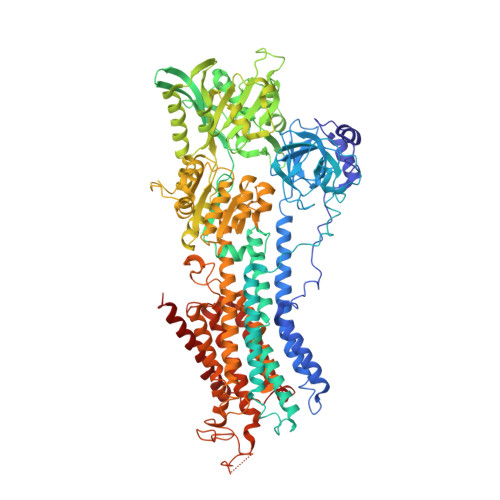
Crystal structure of sarcoplasmic reticulum Ca(2+)-ATPase (SERCA) from bovine muscle.
Sacchetto, R., Bertipaglia, I., Giannetti, S., Cendron, L., Mascarello, F., Damiani, E., Carafoli, E., Zanotti, G.(2012) J Struct Biol 178: 38-44
- PubMed: 22387132
- DOI: https://doi.org/10.1016/j.jsb.2012.02.008
- Primary Citation of Related Structures:
3TLM - PubMed Abstract:
The SERCA pump, a membrane protein of about 110kDa, transports two Ca(2+) ions per ATP hydrolyzed from the cytoplasm to the lumen of the sarcoplasmic reticulum. In muscle cells, its ability to remove Ca(2+) from the cytosol induces relaxation. The transport mechanism employed by the enzyme from rabbit muscle has been extensively studied, and several crystal structures representing different conformational states are available. However, no structure of the pump from other sources is known. In this paper we describe the crystal structure of the bovine enzyme, crystallized in the E1 conformation and determined at 2.9Å resolution. The overall molecular model is very similar to that of the rabbit enzyme, as expected by the high amino acid sequence identity. Nevertheless, the bovine enzyme has reduced catalytic activity with respect to the rabbit enzyme. Subtle structural modifications, in particular in the region of the long loop that protrudes into the SR lumen connecting transmembrane α-helices M7 and M8, may explain the difference.
Organizational Affiliation:
Department of Experimental Veterinary Sciences, Viale dell'Università 16, 35020 Legnaro, Padua, Italy.