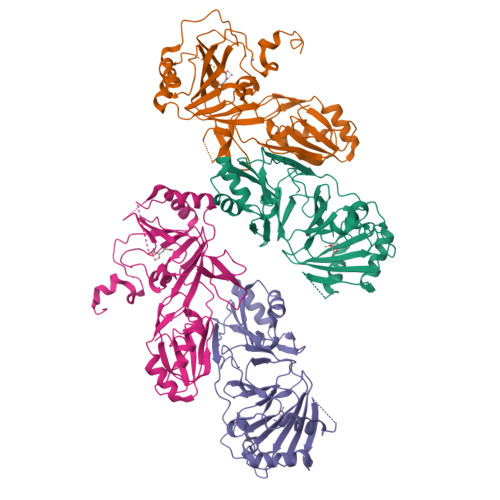
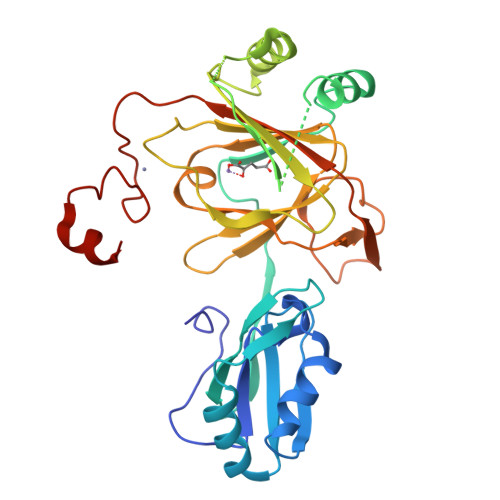
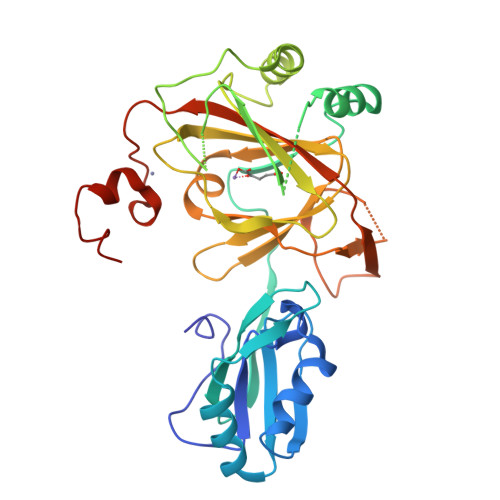
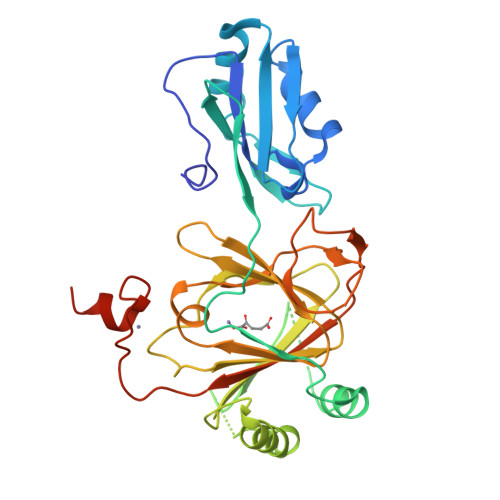
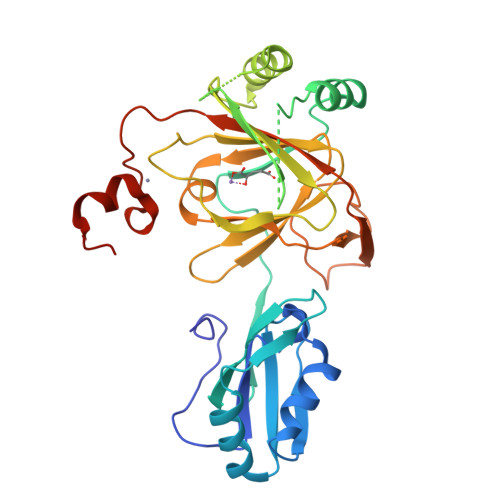
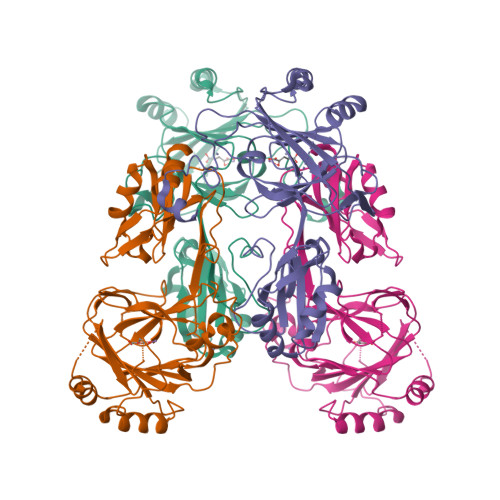
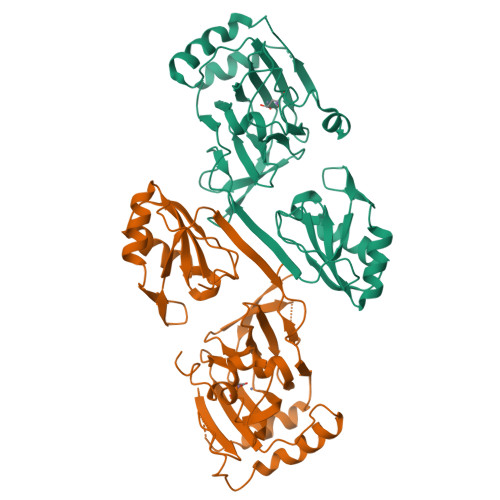
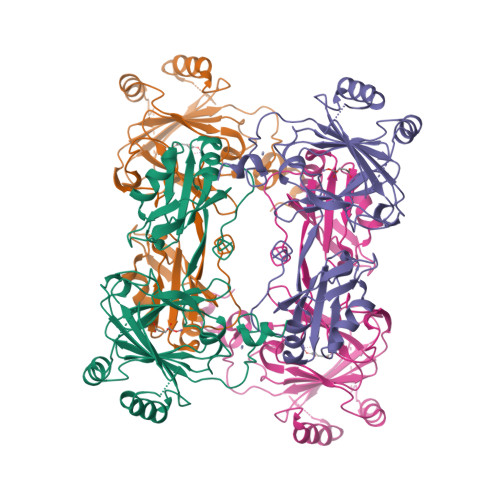
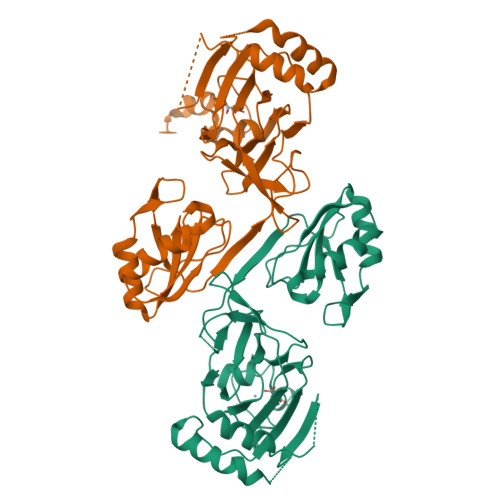
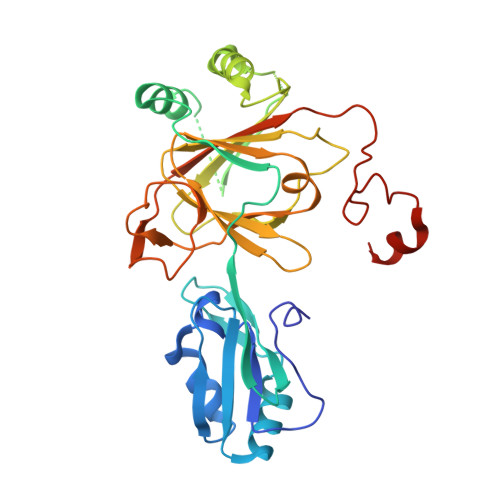
Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
Pastore, C., Topalidou, I., Forouhar, F., Yan, A.C., Levy, M., Hunt, J.F.(2012) J Biological Chem 287: 2130-2143
- PubMed: 22065580
- DOI: https://doi.org/10.1074/jbc.M111.286187
- Primary Citation of Related Structures:
3THP, 3THT - PubMed Abstract:
Humans express nine paralogs of the bacterial DNA repair enzyme AlkB, an iron/2-oxoglutarate-dependent dioxygenase that reverses alkylation damage to nucleobases. The biochemical and physiological roles of these paralogs remain largely uncharacterized, hampering insight into the evolutionary expansion of the AlkB family. However, AlkB homolog 8 (ABH8), which contains RNA recognition motif (RRM) and methyltransferase domains flanking its AlkB domain, recently was demonstrated to hypermodify the anticodon loops in some tRNAs. To deepen understanding of this activity, we performed physiological and biophysical studies of ABH8. Using GFP fusions, we demonstrate that expression of the Caenorhabditis elegans ABH8 ortholog is widespread in larvae but restricted to a small number of neurons in adults, suggesting that its function becomes more specialized during development. In vitro RNA binding studies on several human ABH8 constructs indicate that binding affinity is enhanced by a basic α-helix at the N terminus of the RRM domain. The 3.0-Å-resolution crystal structure of a construct comprising the RRM and AlkB domains shows disordered loops flanking the active site in the AlkB domain and a unique structural Zn(II)-binding site at its C terminus. Although the catalytic iron center is exposed to solvent, the 2-oxoglutarate co-substrate likely adopts an inactive conformation in the absence of tRNA substrate, which probably inhibits uncoupled free radical generation. A conformational change in the active site coupled to a disorder-to-order transition in the flanking protein segments likely controls ABH8 catalytic activity and tRNA binding specificity. These results provide insight into the functional and structural adaptations underlying evolutionary diversification of AlkB domains.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, New York 10027, USA.