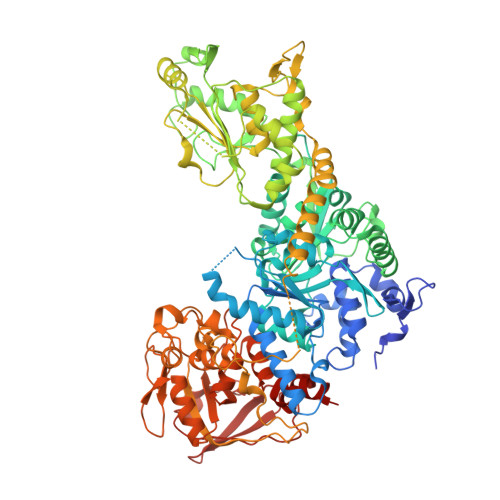
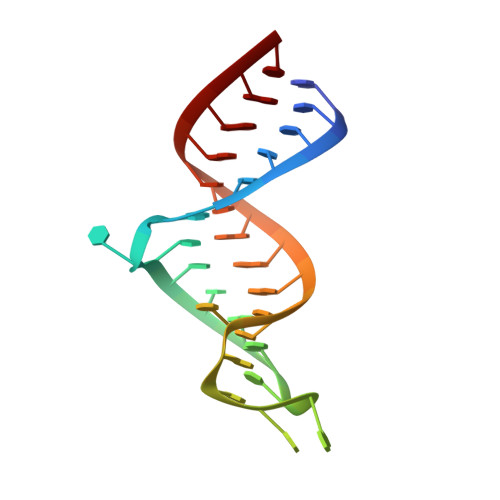
Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA.
Walden, W.E., Selezneva, A.I., Dupuy, J., Volbeda, A., Fontecilla-Camps, J.C., Theil, E.C., Volz, K.(2006) Science 314: 1903-1908
- PubMed: 17185597
- DOI: https://doi.org/10.1126/science.1133116
- Primary Citation of Related Structures:
3SNP - PubMed Abstract:
Iron regulatory protein 1 (IRP1) binds iron-responsive elements (IREs) in messenger RNAs (mRNAs), to repress translation or degradation, or binds an iron-sulfur cluster, to become a cytosolic aconitase enzyme. The 2.8 angstrom resolution crystal structure of the IRP1:ferritin H IRE complex shows an open protein conformation compared with that of cytosolic aconitase. The extended, L-shaped IRP1 molecule embraces the IRE stem-loop through interactions at two sites separated by approximately 30 angstroms, each involving about a dozen protein:RNA bonds. Extensive conformational changes related to binding the IRE or an iron-sulfur cluster explain the alternate functions of IRP1 as an mRNA regulator or enzyme.
- Department of Microbiology and Immunology, University of Illinois at Chicago, Chicago, IL 60612-7344, USA.
Organizational Affiliation: