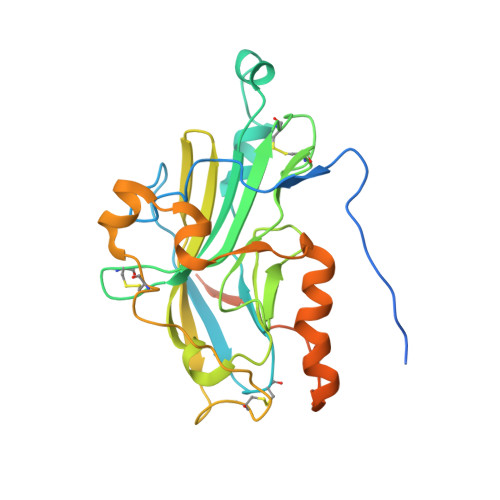
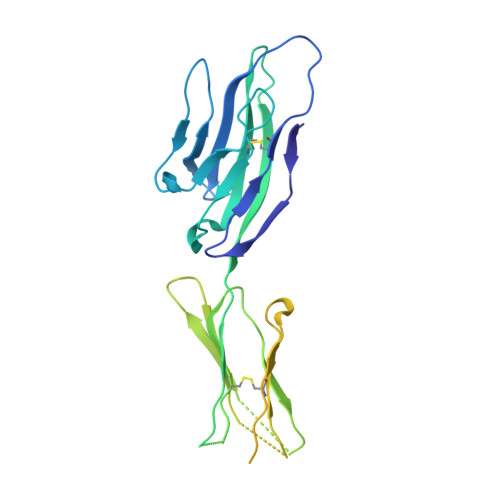
Structure of herpes simplex virus glycoprotein d bound to the human receptor nectin-1.
Di Giovine, P., Settembre, E.C., Bhargava, A.K., Luftig, M.A., Lou, H., Cohen, G.H., Eisenberg, R.J., Krummenacher, C., Carfi, A.(2011) PLoS Pathog 7: e1002277-e1002277
- PubMed: 21980294
- DOI: https://doi.org/10.1371/journal.ppat.1002277
- Primary Citation of Related Structures:
3SKU - PubMed Abstract:
Binding of herpes simplex virus (HSV) glycoprotein D (gD) to a cell surface receptor is required to trigger membrane fusion during entry into host cells. Nectin-1 is a cell adhesion molecule and the main HSV receptor in neurons and epithelial cells. We report the structure of gD bound to nectin-1 determined by x-ray crystallography to 4.0 Å resolution. The structure reveals that the nectin-1 binding site on gD differs from the binding site of the HVEM receptor. A surface on the first Ig-domain of nectin-1, which mediates homophilic interactions of Ig-like cell adhesion molecules, buries an area composed by residues from both the gD N- and C-terminal extensions. Phenylalanine 129, at the tip of the loop connecting β-strands F and G of nectin-1, protrudes into a groove on gD, which is otherwise occupied by C-terminal residues in the unliganded gD and by N-terminal residues in the gD/HVEM complex. Notably, mutation of Phe129 to alanine prevents nectin-1 binding to gD and HSV entry. Together these data are consistent with previous studies showing that gD disrupts the normal nectin-1 homophilic interactions. Furthermore, the structure of the complex supports a model in which gD-receptor binding triggers HSV entry through receptor-mediated displacement of the gD C-terminal region.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, IRBM P. Angeletti, Pomezia, Rome, Italy.