Structural basis for the dual U4 and U4atac snRNA-binding specificity of spliceosomal protein hPrp31.
Liu, S., Ghalei, H., Luhrmann, R., Wahl, M.C.(2011) RNA 17: 1655-1663
- PubMed: 21784869
- DOI: https://doi.org/10.1261/rna.2690611
- Primary Citation of Related Structures:
3SIU, 3SIV - PubMed Abstract:
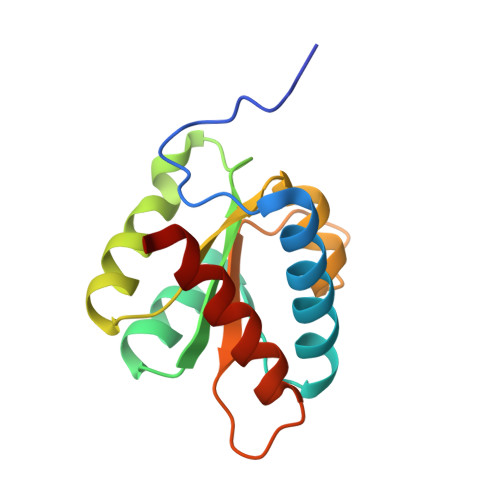
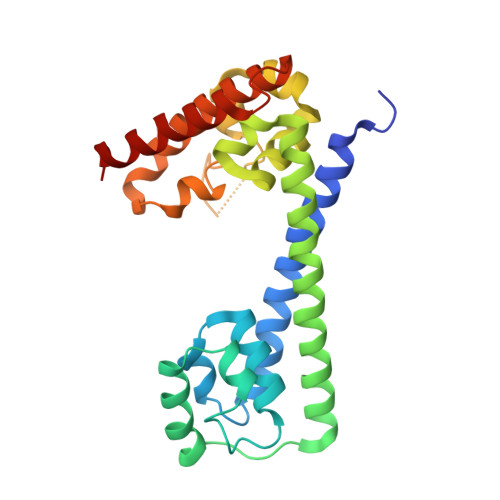
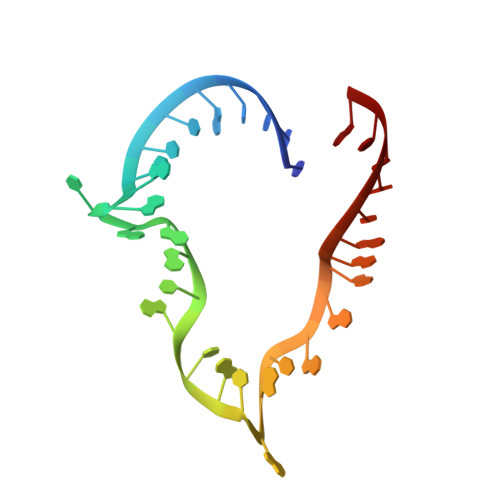
Human proteins 15.5K and hPrp31 are components of the major spliceosomal U4 snRNP and of the minor spliceosomal U4atac snRNP. The two proteins bind to related 5'-stem loops (5'SLs) of the U4 and U4atac snRNAs in a strictly sequential fashion. The primary binding 15.5K protein binds at K-turns that exhibit identical sequences in the two snRNAs. However, RNA sequences contacted by the secondary binding hPrp31 differ in U4 and U4atac snRNAs, and the mechanism by which hPrp31 achieves its dual specificity is presently unknown. We show by crystal structure analysis that the capping pentaloops of the U4 and U4atac 5'SLs adopt different structures in the ternary hPrp31-15.5K-snRNA complexes. In U4atac snRNA, a noncanonical base pair forms across the pentaloop, based on which the RNA establishes more intimate interactions with hPrp31 compared with U4 snRNA. Stacking of hPrp31-His270 on the noncanonical base pair at the base of the U4atac pentaloop recapitulates intramolecular stabilizing principles known from the UUCG and GNRA families of RNA tetraloops. Rational mutagenesis corroborated the importance of the noncanonical base pair and the U4atac-specific hPrp31-RNA interactions for complex stability. The more extensive hPrp31-U4atac snRNA interactions are in line with a higher stability of the U4atac compared with the U4-based ternary complex seen in gel-shift assays, which may explain how U4atac snRNA can compete with the more abundant U4 snRNA for the same protein partners in vivo.
Organizational Affiliation:
Freie Universität Berlin, Fachbereich Biologie/Chemie/Pharmazie, Abteilung Strukturbiochemie, Takustraße 6, D-14195 Berlin, Germany.