How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH.
Eckenroth, B.E., Steere, A.N., Chasteen, N.D., Everse, S.J., Mason, A.B.(2011) Proc Natl Acad Sci U S A 108: 13089-13094
- PubMed: 21788477
- DOI: https://doi.org/10.1073/pnas.1105786108
- Primary Citation of Related Structures:
3S9L, 3S9M, 3S9N - PubMed Abstract:
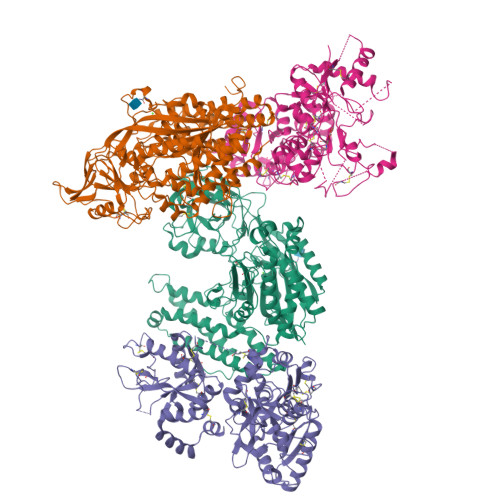
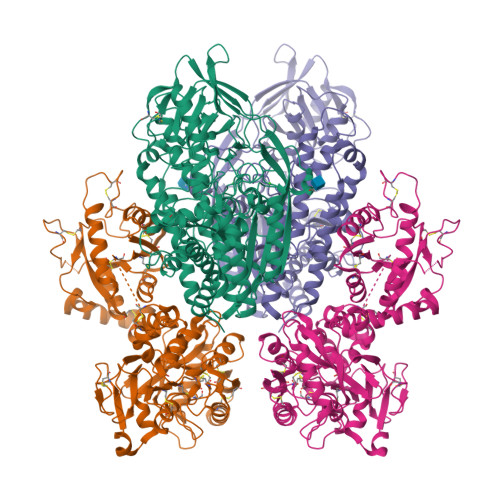
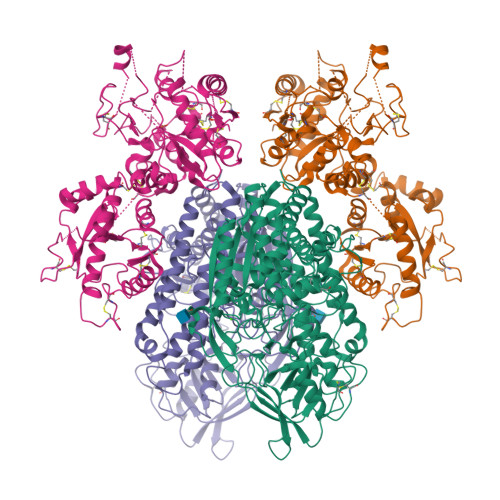
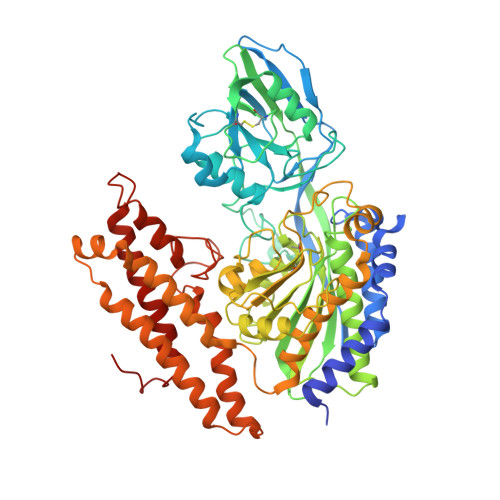
Delivery of iron to cells requires binding of two iron-containing human transferrin (hTF) molecules to the specific homodimeric transferrin receptor (TFR) on the cell surface. Through receptor-mediated endocytosis involving lower pH, salt, and an unidentified chelator, iron is rapidly released from hTF within the endosome. The crystal structure of a monoferric N-lobe hTF/TFR complex (3.22-Å resolution) features two binding motifs in the N lobe and one in the C lobe of hTF. Binding of Fe(N)hTF induces global and site-specific conformational changes within the TFR ectodomain. Specifically, movements at the TFR dimer interface appear to prime the TFR to undergo pH-induced movements that alter the hTF/TFR interaction. Iron release from each lobe then occurs by distinctly different mechanisms: Binding of His349 to the TFR (strengthened by protonation at low pH) controls iron release from the C lobe, whereas displacement of one N-lobe binding motif, in concert with the action of the dilysine trigger, elicits iron release from the N lobe. One binding motif in each lobe remains attached to the same α-helix in the TFR throughout the endocytic cycle. Collectively, the structure elucidates how the TFR accelerates iron release from the C lobe, slows it from the N lobe, and stabilizes binding of apohTF for return to the cell surface. Importantly, this structure provides new targets for mutagenesis studies to further understand and define this system.
Organizational Affiliation:
Department of Biochemistry, University of Vermont, 89 Beaumont Avenue, Burlington, VT 05405, USA.