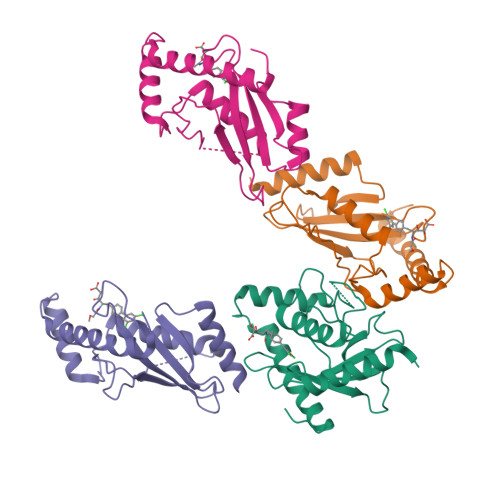
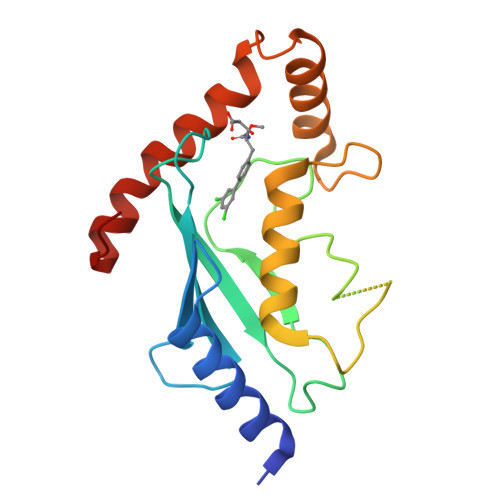
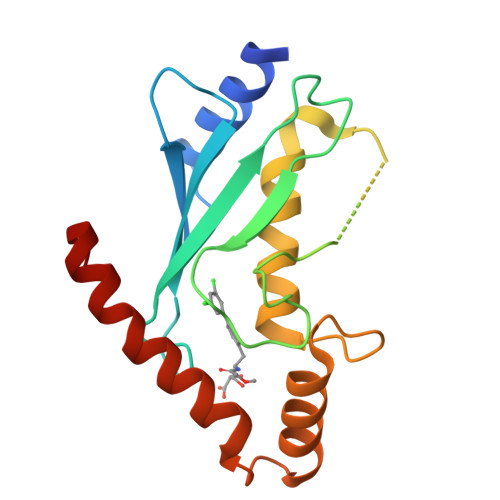
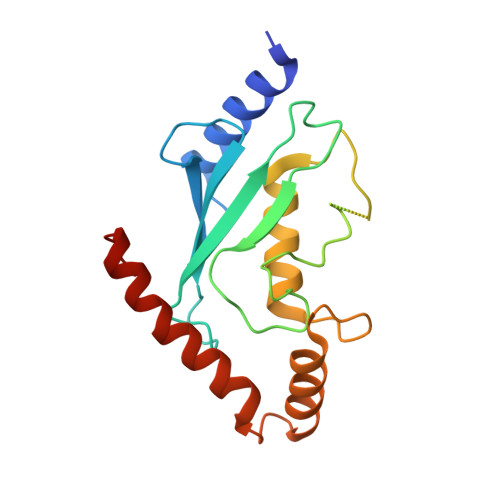
An allosteric inhibitor of the human cdc34 ubiquitin conjugating enzyme
Ceccarelli, D.F., Tang, X., Pelletier, B., Orlicky, S., Xie, W., Plantevin, V., Neculai, D., Chou, Y.C., Ogunjimi, A., Al-Hakim, A., Varelas, X., Koszela, J., Wasney, G.A., Vedadi, M., Dhe-Paganon, S., Cox, S., Xu, S., Lopez-Girona, A., Mercurio, F., Wrana, J., Durocher, D., Meloche, S., Webb, D.R., Tyers, M., Sicheri, F.(2011) Cell 145: 1075-1087
- PubMed: 21683433
- DOI: https://doi.org/10.1016/j.cell.2011.05.039
- Primary Citation of Related Structures:
3RZ3 - PubMed Abstract:
In the ubiquitin-proteasome system (UPS), E2 enzymes mediate the conjugation of ubiquitin to substrates and thereby control protein stability and interactions. The E2 enzyme hCdc34 catalyzes the ubiquitination of hundreds of proteins in conjunction with the cullin-RING (CRL) superfamily of E3 enzymes. We identified a small molecule termed CC0651 that selectively inhibits hCdc34. Structure determination revealed that CC0651 inserts into a cryptic binding pocket on hCdc34 distant from the catalytic site, causing subtle but wholesale displacement of E2 secondary structural elements. CC0651 analogs inhibited proliferation of human cancer cell lines and caused accumulation of the SCF(Skp2) substrate p27(Kip1). CC0651 does not affect hCdc34 interactions with E1 or E3 enzymes or the formation of the ubiquitin thioester but instead interferes with the discharge of ubiquitin to acceptor lysine residues. E2 enzymes are thus susceptible to noncatalytic site inhibition and may represent a viable class of drug target in the UPS.
Organizational Affiliation:
Center for Systems Biology, Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Ontario M5G1X5, Canada.