Crystal structure of enolase PRK14017 from Ralstonia pickettii
Patskovsky, Y., Ramagopal, U.A., Hillerich, B., Seidel, R.D., Zencheck, W.D., Toro, R., Imker, H.J., Gerlt, J.A., Almo, S.C.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative D-galactonate dehydratase | 405 | Ralstonia pickettii 12J | Mutation(s): 0 Gene Names: dgoD, Rpic_2990 EC: 4.2.1.6 | 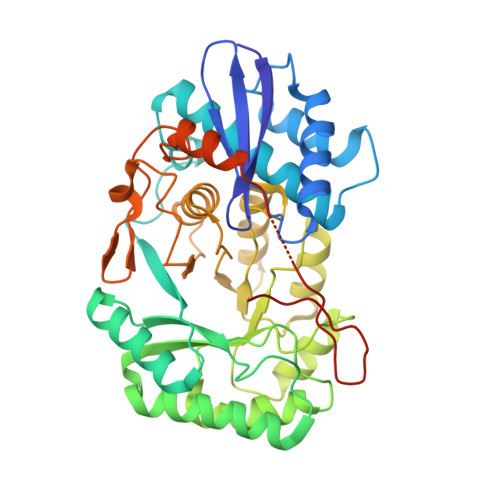 | |
UniProt | |||||
Find proteins for B2UCA8 (Ralstonia pickettii (strain 12J)) Explore B2UCA8 Go to UniProtKB: B2UCA8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B2UCA8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CL Query on CL | C [auth A], E [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | D [auth A], F [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 117.339 | α = 90 |
| b = 117.339 | β = 90 |
| c = 113.685 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHASER | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |