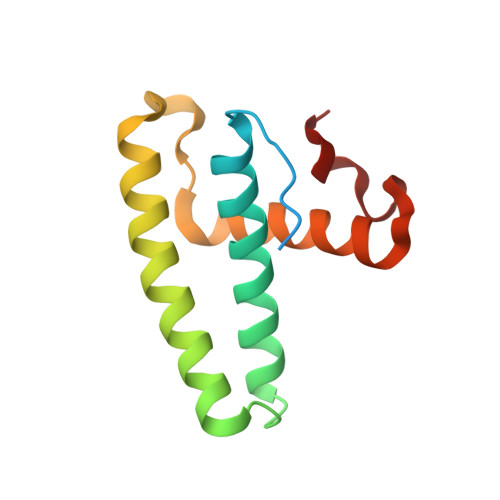
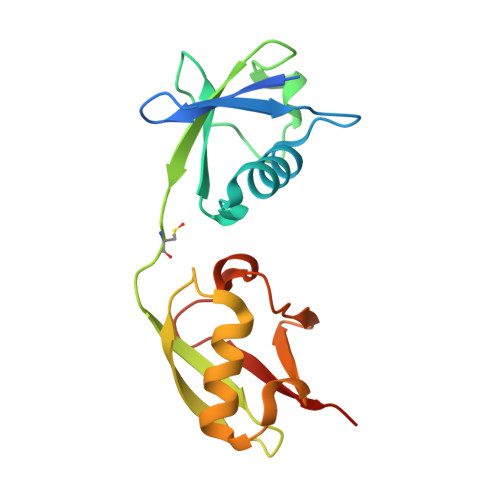
The Structure of the complex of ISG15 and Influenza B virus NS1 Protein: implications for the mechanism of SN1-mediated inhibition of ISG15 conjugation
Guan, R., Ma, L.C., Leonard, P.G., Brendan, A.R., Sridharan, H., Zhao, C., Krug, R.M., Montelione, G.T.To be published.