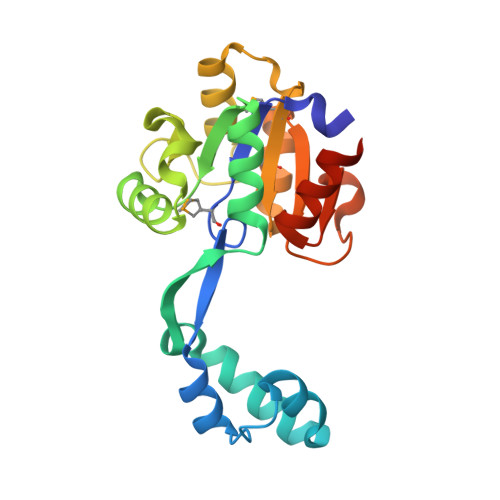
Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg
Vetting, M.W., Patskovsky, Y., Toro, R., Freeman, J., Miller, S., Sauder, J.M., Burley, S.K., Dunaway-Mariano, D., Allen, K.N., Gerlt, J.A., Almo, S.C.To be published.