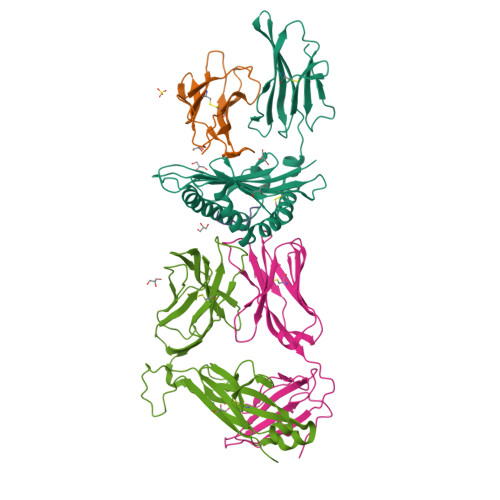
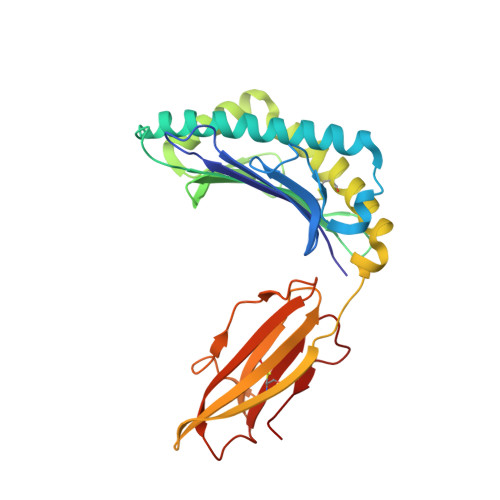
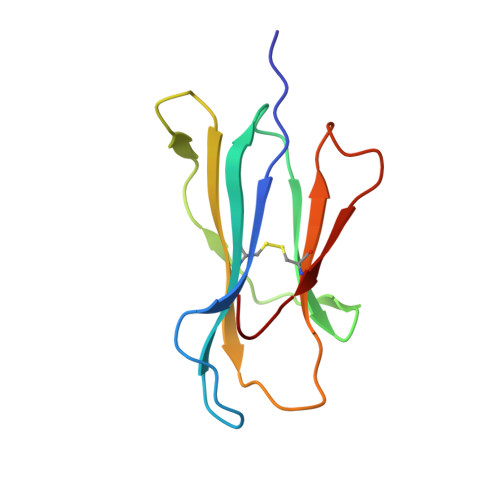
Conformational Melding Permits a Conserved Binding Geometry in TCR Recognition of Foreign and Self Molecular Mimics.
Borbulevych, O.Y., Piepenbrink, K.H., Baker, B.M.(2011) J Immunol 186: 2950-2958
- PubMed: 21282516
- DOI: https://doi.org/10.4049/jimmunol.1003150
- Primary Citation of Related Structures:
3PWJ, 3PWL, 3PWN, 3PWP - PubMed Abstract:
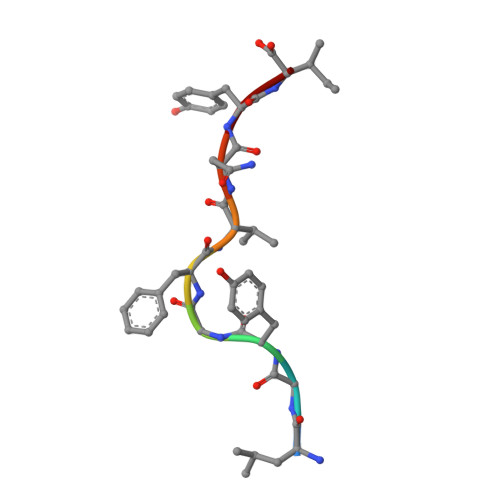
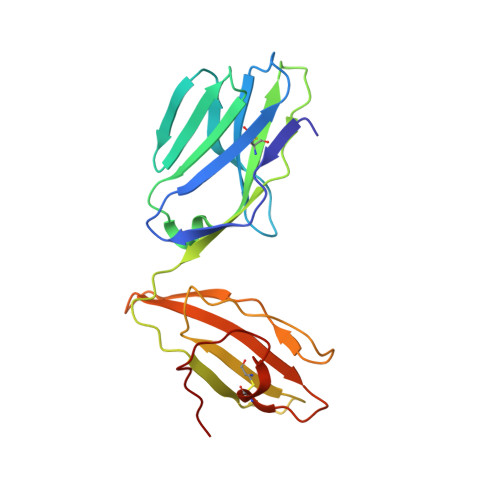
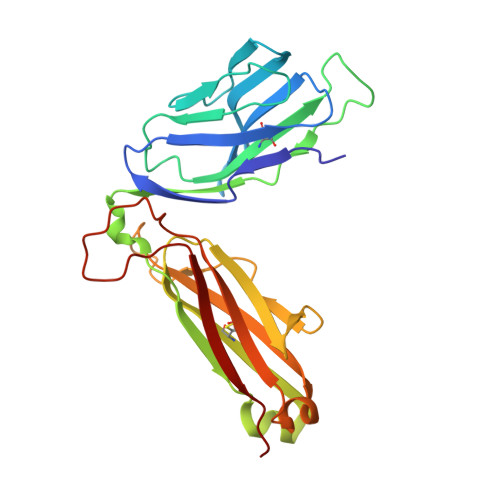
Molecular mimicry between foreign and self Ags is a mechanism of TCR cross-reactivity and is thought to contribute to the development of autoimmunity. The αβ TCR A6 recognizes the foreign Ag Tax from the human T cell leukemia virus-1 when presented by the class I MHC HLA-A2. In a possible link with the autoimmune disease human T cell leukemia virus-1-associated myelopathy/tropical spastic paraparesis, A6 also recognizes a self peptide from the neuronal protein HuD in the context of HLA-A2. We found in our study that the complexes of the HuD and Tax epitopes with HLA-A2 are close but imperfect structural mimics and that in contrast with other recent structures of TCRs with self Ags, A6 engages the HuD Ag with the same traditional binding mode used to engage Tax. Although peptide and MHC conformational changes are needed for recognition of HuD but not Tax and the difference of a single hydroxyl triggers an altered TCR loop conformation, TCR affinity toward HuD is still within the range believed to result in negative selection. Probing further, we found that the HuD-HLA-A2 complex is only weakly stable. Overall, these findings help clarify how molecular mimicry can drive self/nonself cross-reactivity and illustrate how low peptide-MHC stability can permit the survival of T cells expressing self-reactive TCRs that nonetheless bind with a traditional binding mode.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, IN 46556, USA.