X-ray structure of the human calreticulin globular domain reveals a Peptide-binding area and suggests a multi-molecular mechanism
Chouquet, A., Paidassi, H., Ling, W.L., Frachet, P., Houen, G., Arlaud, G.J., Gaboriaud, C.(2011) PLoS One 6: e17886-e17886
- PubMed: 21423620
- DOI: https://doi.org/10.1371/journal.pone.0017886
- Primary Citation of Related Structures:
3POS, 3POW - PubMed Abstract:
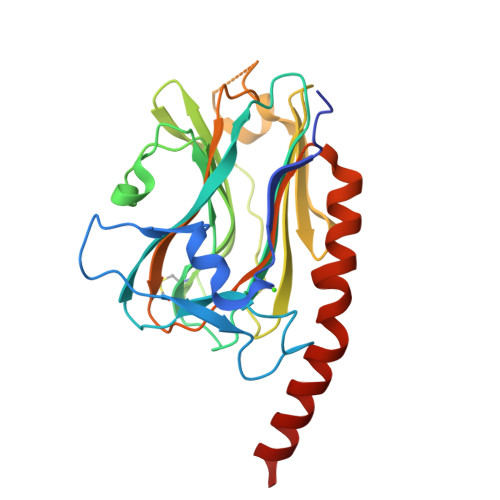
In the endoplasmic reticulum, calreticulin acts as a chaperone and a Ca(2+)-signalling protein. At the cell surface, it mediates numerous important biological effects. The crystal structure of the human calreticulin globular domain was solved at 1.55 Å resolution. Interactions of the flexible N-terminal extension with the edge of the lectin site are consistently observed, revealing a hitherto unidentified peptide-binding site. A calreticulin molecular zipper, observed in all crystal lattices, could further extend this site by creating a binding cavity lined by hydrophobic residues. These data thus provide a first structural insight into the lectin-independent binding properties of calreticulin and suggest new working hypotheses, including that of a multi-molecular mechanism.
Organizational Affiliation:
Institut de Biologie Structurale Jean-Pierre Ebel, CEA, Grenoble, France.