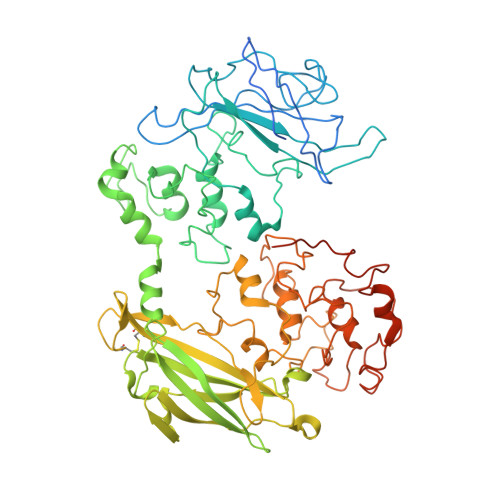
Structure of a bacterial cell surface decaheme electron conduit.
Clarke, T.A., Edwards, M.J., Gates, A.J., Hall, A., White, G.F., Bradley, J., Reardon, C.L., Shi, L., Beliaev, A.S., Marshall, M.J., Wang, Z., Watmough, N.J., Fredrickson, J.K., Zachara, J.M., Butt, J.N., Richardson, D.J.(2011) Proc Natl Acad Sci U S A 108: 9384-9389
- PubMed: 21606337
- DOI: https://doi.org/10.1073/pnas.1017200108
- Primary Citation of Related Structures:
3PMQ - PubMed Abstract:
Some bacterial species are able to utilize extracellular mineral forms of iron and manganese as respiratory electron acceptors. In Shewanella oneidensis this involves decaheme cytochromes that are located on the bacterial cell surface at the termini of trans-outer-membrane electron transfer conduits. The cell surface cytochromes can potentially play multiple roles in mediating electron transfer directly to insoluble electron sinks, catalyzing electron exchange with flavin electron shuttles or participating in extracellular intercytochrome electron exchange along "nanowire" appendages. We present a 3.2-Å crystal structure of one of these decaheme cytochromes, MtrF, that allows the spatial organization of the 10 hemes to be visualized for the first time. The hemes are organized across four domains in a unique crossed conformation, in which a staggered 65-Å octaheme chain transects the length of the protein and is bisected by a planar 45-Å tetraheme chain that connects two extended Greek key split β-barrel domains. The structure provides molecular insight into how reduction of insoluble substrate (e.g., minerals), soluble substrates (e.g., flavins), and cytochrome redox partners might be possible in tandem at different termini of a trifurcated electron transport chain on the cell surface.
- Centre for Molecular and Structural Biochemistry, School of Biological Sciences and School of Chemistry, University of East Anglia, Norwich NR4 7TJ, United Kingdom.
Organizational Affiliation: