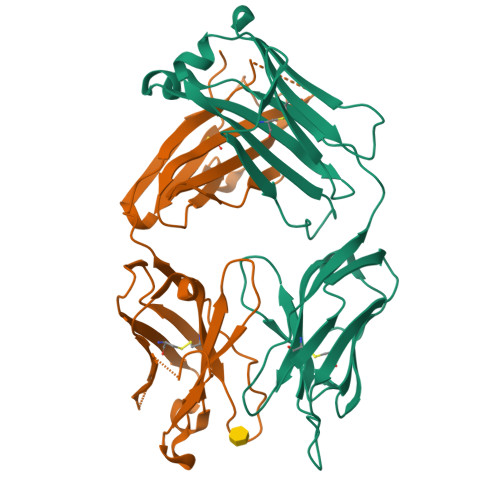
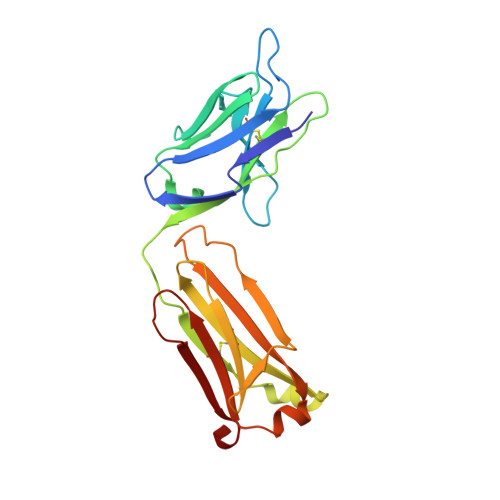
Structural insights into parallel strategies for germline antibody recognition of lipopolysaccharide from Chlamydia.
Evans, D.W., Muller-Loennies, S., Brooks, C.L., Brade, L., Kosma, P., Brade, H., Evans, S.V.(2011) Glycobiology 21: 1049-1059
- PubMed: 21543444
- DOI: https://doi.org/10.1093/glycob/cwr041
- Primary Citation of Related Structures:
3PHO, 3PHQ - PubMed Abstract:
The structure of the antigen-binding fragment from the monoclonal antibody S64-4 in complex with a pentasaccharide bisphosphate fragment from chlamydial lipopolysaccharide has been determined by x-ray diffraction to 2.6 Å resolution. Like the well-characterized antibody S25-2, S64-4 displays a pocket formed by the residues of germline sequence corresponding to the heavy and light chain V gene segments that binds the terminal Kdo residue of the antigen; however, although S64-4 shares the same heavy chain V gene segment as S25-2, it has a different light chain V gene segment. The new light chain V gene segment codes for a combining site that displays greater affinity, different specificity, and allows a novel antigen conformation that brings a greater number of antigen residues into the combining site than possible in S25-2. Further, while antibodies in the S25-2 family use complementarity determining region (CDR) H3 to discriminate among antigens, S64-4 achieves its specificity via the new light chain V gene segment and resulting change in antigen conformation. These structures reveal an intriguing parallel strategy where two different combinations of germline-coded V gene segments can act as starting points for the generation of germline antibodies against chlamydial antigens and show how anti-carbohydrate antibodies can exploit the conformational flexibility of this class of antigens to achieve high affinity and specificity independently of CDR H3.
Organizational Affiliation:
Department of Biochemistry and Microbiology, University of Victoria, PO Box 3055 STN CSC, Victoria, BC, Canada V8P 3P6.