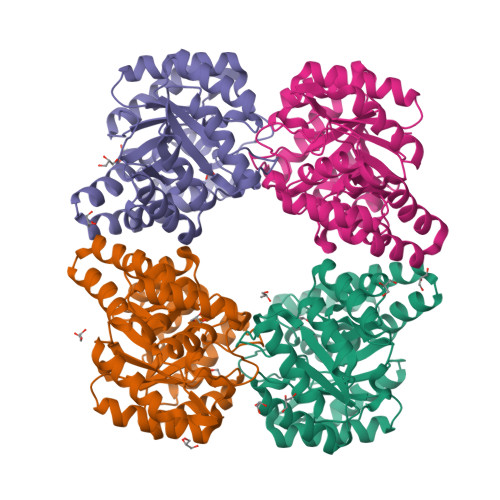
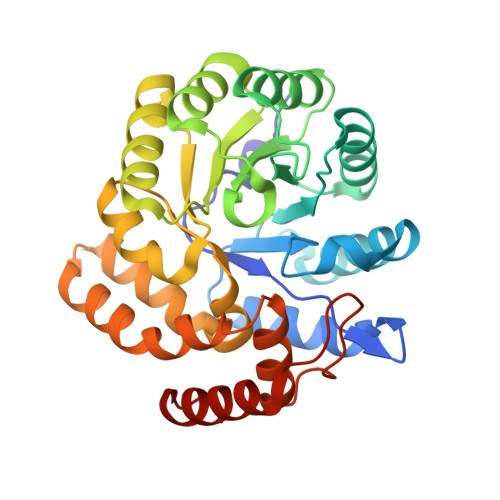
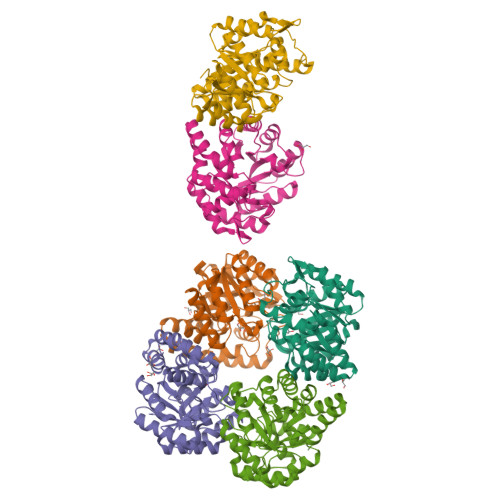Characterization of monomeric dihydrodipicolinate synthase variant reveals the importance of substrate binding in optimizing oligomerization.
Pearce, F.G., Dobson, R.C., Jameson, G.B., Perugini, M.A., Gerrard, J.A.(2011) Biochim Biophys Acta 1814: 1900-1909
- PubMed: 21803176
- DOI: https://doi.org/10.1016/j.bbapap.2011.07.016
- Primary Citation of Related Structures:
3PB0, 3PB2 - PubMed Abstract:
To gain insights into the role of quaternary structure in the TIM-barrel family of enzymes, we introduced mutations to the DHDPS enzyme of Thermotoga maritima, which we have previously shown to be a stable tetramer in solution. These mutations were aimed at reducing the number of salt bridges at one of the two tetramerization interface of the enzyme, which contains many more interactions than the well characterized equivalent interface of the mesophilic Escherichia coli DHDPS enzyme. The resulting variants had altered quaternary structure, as shown by analytical ultracentrifugation, gel filtration liquid chromatography, and small angle X-ray scattering, and X-ray crystallographic studies confirmed that one variant existed as an independent monomer, but with few changes to the secondary and tertiary structure. Reduction of higher order assembly resulted in a loss of thermal stability, as measured by a variety of methods, and impaired catalytic function. Binding of pyruvate increased the oligomeric status of the variants, with a concomitant increase in thermal stability, suggesting a role for substrate binding in optimizing stable, higher order structures. The results of this work show that the salt bridges located at the tetramerization interface of DHDPS play a significant role in maintaining higher order structures, and demonstrate the importance of quaternary structure in determining protein stability and in the optimization of enzyme catalysis.
Organizational Affiliation:
School of Biological Sciences, University of Canterbury, Christchurch, New Zealand. grant.pearce@canterbury.ac.nz